Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA-protein cross-linking - PubMed (original) (raw)
Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA-protein cross-linking
Sander Granneman et al. EMBO J. 2010.
Abstract
Understanding of eukaryotic ribosome synthesis has been slowed by a lack of structural data for the pre-ribosomal particles. We report rRNA-binding sites for six late-acting 40S ribosome synthesis factors, three of which cluster around the 3' end of the 18S rRNA in model 3D structures. Enp1 and Ltv1 were previously implicated in 'beak' structure formation during 40S maturation--and their binding sites indicate direct functions. The kinase Rio2, putative GTPase Tsr1 and dimethylase Dim1 bind sequences involved in tRNA interactions and mRNA decoding, indicating that their presence is incompatible with translation. The Dim1- and Tsr1-binding sites overlap with those of homologous Escherichia coli proteins, revealing conservation in assembly pathways. The primary binding sites for the 18S 3'-endonuclease Nob1 are distinct from its cleavage site and were unaltered by mutation of the catalytic PIN domain. Structure probing indicated that at steady state the cleavage site is likely unbound by Nob1 and flexible in the pre-rRNA. Nob1 binds before pre-rRNA cleavage, and we conclude that structural reorganization is needed to bring together the catalytic PIN domain and its target.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
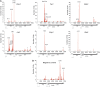
Overview of CRAC results. Shown are the results from independent CRAC experiments performed on pre-40S-associated proteins (A). Results from untagged strains are shown in (B). Sequences were aligned to the rDNA reference sequence using blast and plotted using gnuplot. The locations of mature rRNA sequences, spacers and cleavage site are indicated below the x axis. The y axis shows the total number of times each nucleotide within an RNA fragment was mapped to the rDNA sequence. The location of the peaks in the secondary structure of the rRNA (see Figure 2A, B) is indicated with helix (H) numbers. The asterisks indicate frequent contaminants.
Figure 2
Locations of protein–RNA interaction sites in the 18S rRNA secondary structure. (A) Overview of the yeast 18S rRNA secondary structure (obtained from
http://www.rna.ccbb.utexas.edu/
). The stem including the D-cleavage site was added separately. The 18S rRNA domains are indicated as dashed boxes. Functionally, important elements (central pseudoknot, decoding centre, P-site tRNA interactions, D-cleavage site (coloured red)) are marked with circles. (B) Overview of RNA-binding sites for pre-40S-associated proteins. Specific nucleotides cross-linked to proteins are indicated as coloured circles. For cases where no or few mutations were found in cross-linked RNAs (Tsr1 H26 and H27, Dim1 H28 and the Enp1-binding site in H34), the binding sites are indicated by dashed lines. Dashed boxes indicate predicted ribosomal protein (Rps) interaction sites on the 18S rRNA, based on the locations of homologous T. thermophilus ribosomal proteins in the 30S crystal structure, the yeast 40S structure model and various other studies performed on ribosomal proteins in yeast (Spahn et al, 2001; Brodersen et al, 2002; Antunez de Mayolo and Woolford, 2003; Dresios et al, 2005). Helix numbering was adopted from Brodersen et al (2002). RNA-binding sites for the RNA helicase Prp43 (Bohnsack et al, 2009) are also included.
Figure 3
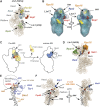
Modelling of protein–RNA interaction sites into 3D structures. (A) Modelling of protein–RNA-binding sites in the 40S structure model (pdb 1s1h; Spahn et al, 2001). Shown is the model structure of the 18S rRNA. Characteristic structural features are indicated as head, beak, neck, shoulder and platform. Enp1- and Ltv1-binding sites are located near the beak formed by H33. Shown in wheat colour is the 18S rRNA tertiary structure superimposed with ribbon structure models for Rps3 (light blue), Rps9 (green) and Rps20 (yellow). H16 and H41A that interact with Ltv1 are shown in black. Enp1 cross-linking sites in H33 are depicted as red dots. (B) Ltv1 may correspond to the extra density near the head and the shoulder in the head domain. The docked molecular model of the yeast 40S subunit is shown for the 40S cryo-EM map (cage) superimposed on the pre-40S cryo-EM map (transparent blue). Helices 16 and 41 surfaces (Ltv1-binding sites) are indicated in yellow, Enp1-binding site surface is indicated in red, Rps3 is indicated in blue and Rps16 is indicated in orange. (C) Model for beak formation in pre-40S complexes. On the schematic representation of pre-40S cryo-EM reconstruction images (Schafer et al, 2006) the predicted location of Rps3 is indicated in blue. In the model, Hrr25-dependent phosphorylation of Rps3 and subsequent dephosphorylation trigger a structural change leading to repositioning of Rps3, releasing of Enp1 and Ltv1 and beak formation (Schafer et al, 2006). Included are the predicted locations of Ltv1 (spanning the head and shoulder domain) and Enp1 (in the centre next to Rps3) in the pre-40S pre-ribosome. (D) Rio2, Ltv1 and Nob1-binding sites cluster around Rps16. The surface density of Rps15 and Rps16 in the head domain of the small subunit is shown in green and orange, respectively. The cross-linking sites for Ltv1 (black), Nob1 (red) and Rio2 (blue) are indicated as dots. (E) Tsr1 and Dim1 interact with rRNA near the decoding centre. The Tsr1- and Dim1-binding sites were modelled in the T. thermophilus 30S crystal structure with bound mRNA (PDB 2wgd) (Voorhees et al, 2009). Shown are helices 44, 45 and the RNA segments containing the Tsr1- and Dim1-binding sites (helices 11,26,27,28). The decoding centre is roughly marked with a dashed circle. (F) Nob1 binds the head domain in close proximity to the 3′ end of the 18S rRNA. Shown is the tertiary structure model of the 18S rRNA. Nob1 and Prp43 cross-linking sites are indicated as coloured dots. Helix 44 is indicated in black. The arrows indicate the distance (33 Å) between the Nob1 cross-linking site and the modelled 3′-end of the 18S rRNA.
Figure 4
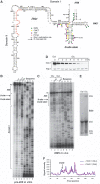
At steady state the stem structure containing the D-cleavage site is highly flexible and unbound by proteins. (A) Overview of the chemical foot-printing results on a secondary structure of ITS1, including the 5′ end of the 18S rRNA, adopted from Yeh et al (1990). Red nucleotides indicate the location of the reverse transcriptase primer. Adenosines in H45 dimethylated by Dim1 in vivo are indicated with ‘-m'. Green circles indicate the nucleotide modified by DMS. Yellow and purple dots indicate the CMCT and 1M7-modified nucleotides, respectively. (B) The D-site region in purified 20S pre-rRNA is flexible and unbound by proteins. DMS, CMCT and 1M7 modification of RNA was performed at 30°C on purified pre-40S complexes isolated using plasmid-expressed HTP-tagged Nob1 as bait. All chemicals primarily modified nucleotides in the D-site stem region, indicating it is flexible or single stranded, whereas other regions predicted to be double stranded (i.e. H45 and Domain I) were largely protected from chemical modification. Adenosines in H45 dimethylated by Dim1 in vivo are indicated with ‘-m'. (C–F) The D-site helix is highly flexible and likely unbound by Nob1 in vivo. In vivo DMS was performed at 30°C on total RNA purified from GAL::3HA-nob1 cells (E) grown in glucose for 8 h (C, lanes 1 and 2) or galactose (C, lanes 3 and 4). Depletion of Nob1 was confirmed by western blotting (D) using an HRP-conjugated anti-HA antibody (Santa Cruz) and accumulation of 20S pre-rRNA was detected by ethidium bromide staining of total RNA in an agarose gel. Pgk1 antibodies (Santa Cruz) were used to confirm loading of equal amounts proteins on each lane (D). After primer extension, radiolabelled cDNAs were resolved on 12% polyacrylamide/7 M urea gels and visualized by autoradiography. Adenosines in H45 dimethylated by Dim1 in vivo are indicated with ‘-m'. Quantification of chemical probing data (F) was performed as described in the main text.
Figure 5
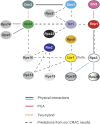
Overview of known and predicted protein–protein interactions in pre-40S complexes. The interaction map depicts interactions between the various assembly factors and ribosomal proteins in pre-40S complexes. Black lines, physical interactions among proteins shown to be part of subcomplexes or interacting as recombinant proteins (Krogan et al, 2004; Vanrobays et al, 2004; Schafer et al, 2006; Tarassov et al, 2008; Lamanna and Karbstein, 2009). Dashed red lines, yeast two-hybrid interactions (Ito et al, 2001; Tone and Toh, 2002). Red lines, interactions from protein–fragment complementation assays (PCA), which detect proteins within 80 Å distance from each other (Tarassov et al, 2008). Dashed black lines, protein–protein interactions predicted from our CRAC data. Blue lines, physical interactions among bacterial homologues of the indicated proteins (Brodersen et al, 2002).
Similar articles
- Rio1 mediates ATP-dependent final maturation of 40S ribosomal subunits.
Turowski TW, Lebaron S, Zhang E, Peil L, Dudnakova T, Petfalski E, Granneman S, Rappsilber J, Tollervey D. Turowski TW, et al. Nucleic Acids Res. 2014 Oct 29;42(19):12189-99. doi: 10.1093/nar/gku878. Epub 2014 Oct 7. Nucleic Acids Res. 2014. PMID: 25294836 Free PMC article. - Cryo-EM structure of a late pre-40S ribosomal subunit from Saccharomyces cerevisiae.
Heuer A, Thomson E, Schmidt C, Berninghausen O, Becker T, Hurt E, Beckmann R. Heuer A, et al. Elife. 2017 Nov 20;6:e30189. doi: 10.7554/eLife.30189. Elife. 2017. PMID: 29155690 Free PMC article. - Nob1 binds the single-stranded cleavage site D at the 3'-end of 18S rRNA with its PIN domain.
Lamanna AC, Karbstein K. Lamanna AC, et al. Proc Natl Acad Sci U S A. 2009 Aug 25;106(34):14259-64. doi: 10.1073/pnas.0905403106. Epub 2009 Aug 14. Proc Natl Acad Sci U S A. 2009. PMID: 19706509 Free PMC article. - Inside the 40S ribosome assembly machinery.
Karbstein K. Karbstein K. Curr Opin Chem Biol. 2011 Oct;15(5):657-63. doi: 10.1016/j.cbpa.2011.07.023. Epub 2011 Aug 20. Curr Opin Chem Biol. 2011. PMID: 21862385 Free PMC article. Review. - Maturation of pre-40S particles in yeast and humans.
Cerezo E, Plisson-Chastang C, Henras AK, Lebaron S, Gleizes PE, O'Donohue MF, Romeo Y, Henry Y. Cerezo E, et al. Wiley Interdiscip Rev RNA. 2019 Jan;10(1):e1516. doi: 10.1002/wrna.1516. Epub 2018 Nov 8. Wiley Interdiscip Rev RNA. 2019. PMID: 30406965 Review.
Cited by
- Flatworms have lost the right open reading frame kinase 3 gene during evolution.
Breugelmans B, Ansell BR, Young ND, Amani P, Stroehlein AJ, Sternberg PW, Jex AR, Boag PR, Hofmann A, Gasser RB. Breugelmans B, et al. Sci Rep. 2015 May 15;5:9417. doi: 10.1038/srep09417. Sci Rep. 2015. PMID: 25976756 Free PMC article. - The DEAH-box helicase Dhr1 dissociates U3 from the pre-rRNA to promote formation of the central pseudoknot.
Sardana R, Liu X, Granneman S, Zhu J, Gill M, Papoulas O, Marcotte EM, Tollervey D, Correll CC, Johnson AW. Sardana R, et al. PLoS Biol. 2015 Feb 24;13(2):e1002083. doi: 10.1371/journal.pbio.1002083. eCollection 2015 Feb. PLoS Biol. 2015. PMID: 25710520 Free PMC article. - Understanding splicing regulation through RNA splicing maps.
Witten JT, Ule J. Witten JT, et al. Trends Genet. 2011 Mar;27(3):89-97. doi: 10.1016/j.tig.2010.12.001. Epub 2011 Jan 12. Trends Genet. 2011. PMID: 21232811 Free PMC article. Review. - Final pre-40S maturation depends on the functional integrity of the 60S subunit ribosomal protein L3.
García-Gómez JJ, Fernández-Pevida A, Lebaron S, Rosado IV, Tollervey D, Kressler D, de la Cruz J. García-Gómez JJ, et al. PLoS Genet. 2014 Mar 6;10(3):e1004205. doi: 10.1371/journal.pgen.1004205. eCollection 2014 Mar. PLoS Genet. 2014. PMID: 24603549 Free PMC article. - Rio1 mediates ATP-dependent final maturation of 40S ribosomal subunits.
Turowski TW, Lebaron S, Zhang E, Peil L, Dudnakova T, Petfalski E, Granneman S, Rappsilber J, Tollervey D. Turowski TW, et al. Nucleic Acids Res. 2014 Oct 29;42(19):12189-99. doi: 10.1093/nar/gku878. Epub 2014 Oct 7. Nucleic Acids Res. 2014. PMID: 25294836 Free PMC article.
References
- Antunez de Mayolo P, Woolford JL Jr. (2003) Interactions of yeast ribosomal protein rpS14 with RNA. J Mol Biol 133: 697–709 - PubMed
- Ares M Jr, Igel AH (1990) Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes Dev 4: 2132–2145 - PubMed
- Brodersen DE, Clemons WM Jr, Carter AP, Wimberly BT, Ramakrishnan V (2002) Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J Mol Biol 316: 725–768 - PubMed
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G0900740/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- BB/D019621/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases