PERK promotes cancer cell proliferation and tumor growth by limiting oxidative DNA damage - PubMed (original) (raw)
PERK promotes cancer cell proliferation and tumor growth by limiting oxidative DNA damage
E Bobrovnikova-Marjon et al. Oncogene. 2010.
Abstract
To proliferate and expand in an environment with limited nutrients, cancer cells co-opt cellular regulatory pathways that facilitate adaptation and thereby maintain tumor growth and survival potential. The endoplasmic reticulum (ER) is uniquely positioned to sense nutrient deprivation stress and subsequently engage signaling pathways that promote adaptive strategies. As such, components of the ER stress-signaling pathway represent potential antineoplastic targets. However, recent investigations into the role of the ER resident protein kinase, RNA-dependent protein kinase (PKR)-like ER kinase (PERK) have paradoxically suggested both pro- and anti-tumorigenic properties. We have used animal models of mammary carcinoma to interrogate the contribution of PERK in the neoplastic process. The ablation of PERK in tumor cells resulted in impaired regeneration of intracellular antioxidants and accumulation of reactive oxygen species triggering oxidative DNA damage. Ultimately, PERK deficiency impeded progression through the cell cycle because of the activation of the DNA damage checkpoint. Our data reveal that PERK-dependent signaling is used during both tumor initiation and expansion to maintain redox homeostasis, thereby facilitating tumor growth.
Figures
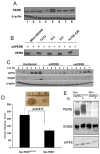
Figure 1. PERK expression is maintained in cancer cells wherein it regulates tumor expansion in vivo
(A) PERK protein levels were measured by immunoprecipitation (IP) followed by Western blot analysis in the following cell lines: MCF10A (1), MCF7 (2), T47D (3), MDA-MB231 (4), MDA-MB468 (5), TE3 (6), TE7 (7), KYSE 520 (8). (B) PERK protein levels following shRNA targeting of PERK. (C) Parental MDA-MB468 cell line, shPERK-transduced cells (shPERK), and shPERK-transduced cells reconstituted with mouse Myc-PERK (+mPERK) were treated with 2μg/ml tunicamycin for the indicated intervals. Western analysis for ATF4, CHOP, or β-actin. (D) Volume of orthotopic tumors formed from the mouse mammary tumor-derived cells transduced in vitro with empty vector virus (Neu/PERKloxP/loxP) or Cre-expressing retrovirus (Neu/PERKΔ/Δ) 28 days post-transplant (n=4). Representative image of tumors are provided. All p-values determined by Student t-test. (E) Western analysis of transgenic ErbB2 and PERK expression following infection of mouse mammary tumor-derived cells with control (Neu/PERKloxP/loxP) or Cre-expressing retrovirus (Neu/PERKΔ/Δ). Thapsigargin treatment (50nM, 1h) was used to demonstrate that PERK is functional.
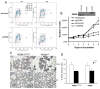
Figure 2. PERK knockdown triggers a G2/M delay
(A) MDA-MB468 cells were infected with control shRNA or anti-PERK shRNA for the indicated intervals. Cells were pulsed with BrdU 45 min prior to harvest for FACS analysis. (B) Kinetics of growth of the MDA-MB468 parental cell line, control shRNA-(shControl) or shPERK-transduced cells (shPERK), and shPERK-transduced cells reconstituted with mouse Myc-PERK (+mPERK). PERK protein levels following expression of shRNA targeting human PERK and reconstitution with mouse Myc-PERK are shown. (C) Proliferation rates in mammary gland sections from control (PERKloxP/loxP) and mammary gland-specific PERK knockout mice (PERKΔ/Δ) on pregnancy day 16 (P16) and lactation day 3 (L3) were determined by immunohistochemistry for BrdU (animals were injected with BrdU 1 h prior to being euthanized). (D) Quantification of BrdU-positive cells from (C) is shown; error bars indicate S.D. among 3 animals, 5 acini were counted per animal.
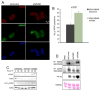
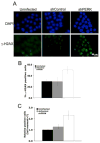
Figure 3. PERK knockdown triggers DNA damage response signaling pathway
(A) Immunofluorescence staining for DNA damage-induced foci containing phospho-ATM and phospho-Chk2 following acute PERK knockdown (72 h after infection) in MDA-MB468 cells. (B) Quantification of phospho-ATM positive cells (>3 foci) is shown; error bars indicate S.D. from 3 slides, 5 fields were counted per slide. p-value was determined by Student t-test. (C) Western analysis of DNA damage response-associated markers following PERK knockdown. (D) IP/kinase assays assessing CDK2-dependent phosphorylation of histone H1 (bottom panel). CDK2 complexes were immunoprecipitated from MDA-MB 468 cells treated as indicated. PERK levels were assessed by IP/immunoblot and CDK2 recovery in precipitates was assessed by CDK2 immunoblot (middle panel).
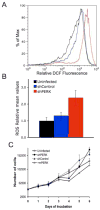
Figure 4. Increased levels of Reactive Oxygen Species (ROS) in PERK knockdown cells contribute to reduced kinetics of cell growth
(A) DCF fluorescence measured by FACS using CM-H2DCFDA dye. (B) Mean values of ROS determined from three independent experiments. Uninfected (black), stably infected with empty vector (blue) or shPERK (red) in both (A) and (B). (C) Growth analysis of MDA-MB468 parental cell line, cells transduced with control shRNA, shPERK, or shPERK and reconstituted with mouse Myc-PERK in the presence of ROS scavenger N-acetylcysteine (NAC).
Figure 5. ROS accumulation triggers oxidative DNA lesions in PERK-deficient breast cancer cells and tumors
(A) Detection of 8-oxoguanine oxidized DNA adduct (8-OxoG) using a FITC conjugated 8-OxoG binding peptide in parental MDA-MB468 cells, MDA-MB468 cells transduced with control shRNA, shPERK, or shPERK and reconstituted with mouse Myc-PERK. (B) Quantification of 8-oxoG positive cells from 3 independent experiments. (C) 8-OxoG in paraffin sections from tumors formed by parental MDA-MB468 cells, MDA-MB468 cells transduced with control shRNA, or shPERK. (D) Quantification of 8-OxoG positive cells shown in (C) is provided and error bars indicate S.D. from 4 animals. (E) Detection of 8-OxoG in paraffin sections from orthotopic tumors formed by mouse mammary tumor-derived cells transduced with empty vector (Neu/PERKloxP/loxP) or retrovirus expressing Cre recombinase (Neu/PERKΔ/Δ). (F) Quantification of 8-OxoG positive cells from (E); error bars indicate S.D. from 4 animals. All p-values determined by Student t-test.
Figure 6. ROS accumulation triggers DNA double strand breaks in PERK-deficient breast cancer cells and tumors
(A) Immunofluorescent staining of γ-H2AX foci in control or PERK knockdown MDA-MB468 cells. (B) Quantification of γ-H2AX positive (>5 foci) cells under standard tissue culture conditions. Error bars represent S.D. from 3 independent experiments performed in triplicate. (C) Quantification of the COMET tail moment in control or PERK knockdown cells under standard tissue culture conditions. Error bars indicate S.D. from 3 experiments.
Figure 7. Reduced activity of Nrf2 causes increased oxidative stress in PERK knockdown cells
(A) Quantitative real time PCR analysis of Nrf2 target genes NQO1 and GCLC in the indicated cell lines asynchronously proliferating under standard conditions. (B) Purified recombinant Nrf2-Neh2 domain of WT, T80A, S40A or T80A/S40A, was incubated with purified recombinant ΔN-PERK in the in vitro kinase assay. Phosphorylated Nrf2-Neh2 was detected by autoradiography (upper panel). (C) 293T cells were transfected with WT Nrf2 or Nrf2-T80A. 24 hours after transfection, cells were left untreated (C) or treated with tunicamycin (Tu) for 2 hours followed by immunoprecipitation with anti-Nrf2 antibody. Threonine phosphorylation was detected using a phospho-Thr reactive antibody. Nrf2 in the IP and the whole cell lysate (WCL) was detected with Nrf2 specific antibody. (D) Proliferation of the indicated cell lines was assessed by a 6-day growth curve under standard tissue culture conditions as described in materials and methods. PERK levels were detected by IP/Western blot analysis. (E) Oxidized guanine in damaged DNA was detected by a FITC-conjugated 8-OxoG binding peptide in PERK knockdown cells infected with pBabe control vector (shPERK) or Nrf2-HA (shPERK Nrf2-HA). Quantification of 8-OxoG positive cells is provided. Error bars represent S.D. from 3 experiments. (F) 8-OxoG was detected in PERK knockdown cells transfected with scramble siRNA (Scrm), or keap1 siRNA (sikeap1). Error bars in graphs represent S.D. from 3 experiments. Western blot panels demonstrate levels of Nrf2-HA and Keap1 in PERK knockdown cells.
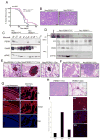
Figure 8. PERK loss attenuates MMTV-_Neu_-driven mammary tumorigenesis in mice, but promotes spontaneous mammary tumor formation in aged mammary gland-specific PERK knockout mice
(A) Kaplan-Meier analysis of tumor-free survival for cohorts of MMTV-Neu/PERKloxP/loxP (n=21) and MMTV-Neu/PERKΔ/Δ Δ/Δ (n=27) mice. (B) Hematoxylin and eosin staining demonstrating histology of control (Neu/PERKloxP/loxP) and PERK knockout (Neu PERKΔ/Δ) mammary gland tumors. (C) Western analysis for PERK, ErbB2, and eIF4E levels on whole protein extracts from control (Neu/PERKloxP/loxP) and PERK knockout (Neu/PERKΔ/Δ) mammary gland tumors or mammary gland from lactating PERKloxP/loxP dam (L10). (D) Nrf2 was precipitated from tumor lysates prepared from MMTV-Neu/PERKΔ/Δ or control mice and blotted for phospho-Thr and Nrf2. PERK expression was determined by immunoblot. (E) PERK excision delays development of Neu-driven hyperplastic lesions. Representative mammary glands from 9- to 14-months old control (Neu/PERKloxP/loxP) and PERK knockout (Neu/PERKΔ/Δ) mice revealing pre-malignant lesions are shown. (F) Hematoxylin and eosin staining on lungs from control (Neu/PERKloxP/loxP, n=24) and PERK knockout (Neu/PERKΔ/Δ, n=27) mice revealing metastatic lesions. (G) Troma-1 (cytokeratin-8) staining on lung specimens containing metastatic foci. LT=lung tissue; Met=metastasis; OL=overlay. (H) Hematoxylin and eosin staining for tumor histology and whole mount of hyperplastic lesions in mammary glands of PERKΔ/Δ aged females. (I) qRT-PCR for ErbB2 on genomic DNA from PERKΔ/Δ tumors and FISH analysis on paraffin sections from the same animals. Levels of ErbB2 in tumors were compared to matched spleen tissues.
Similar articles
- De-differentiation confers multidrug resistance via noncanonical PERK-Nrf2 signaling.
Del Vecchio CA, Feng Y, Sokol ES, Tillman EJ, Sanduja S, Reinhardt F, Gupta PB. Del Vecchio CA, et al. PLoS Biol. 2014 Sep 9;12(9):e1001945. doi: 10.1371/journal.pbio.1001945. eCollection 2014 Sep. PLoS Biol. 2014. PMID: 25203443 Free PMC article. - TBL2 is a novel PERK-binding protein that modulates stress-signaling and cell survival during endoplasmic reticulum stress.
Tsukumo Y, Tsukahara S, Furuno A, Iemura S, Natsume T, Tomida A. Tsukumo Y, et al. PLoS One. 2014 Nov 13;9(11):e112761. doi: 10.1371/journal.pone.0112761. eCollection 2014. PLoS One. 2014. PMID: 25393282 Free PMC article. - Imiquimod-induced autophagy is regulated by ER stress-mediated PKR activation in cancer cells.
Chang SH, Huang SW, Wang ST, Chung KC, Hsieh CW, Kao JK, Chen YJ, Wu CY, Shieh JJ. Chang SH, et al. J Dermatol Sci. 2017 Aug;87(2):138-148. doi: 10.1016/j.jdermsci.2017.04.011. Epub 2017 May 2. J Dermatol Sci. 2017. PMID: 28601430 - Tumor progression and the different faces of the PERK kinase.
Pytel D, Majsterek I, Diehl JA. Pytel D, et al. Oncogene. 2016 Mar 10;35(10):1207-15. doi: 10.1038/onc.2015.178. Epub 2015 Jun 1. Oncogene. 2016. PMID: 26028033 Free PMC article. Review. - The Role of the PERK/eIF2α/ATF4/CHOP Signaling Pathway in Tumor Progression During Endoplasmic Reticulum Stress.
Rozpedek W, Pytel D, Mucha B, Leszczynska H, Diehl JA, Majsterek I. Rozpedek W, et al. Curr Mol Med. 2016;16(6):533-44. doi: 10.2174/1566524016666160523143937. Curr Mol Med. 2016. PMID: 27211800 Free PMC article. Review.
Cited by
- Endoplasmic reticulum and the unfolded protein response: dynamics and metabolic integration.
Bravo R, Parra V, Gatica D, Rodriguez AE, Torrealba N, Paredes F, Wang ZV, Zorzano A, Hill JA, Jaimovich E, Quest AF, Lavandero S. Bravo R, et al. Int Rev Cell Mol Biol. 2013;301:215-90. doi: 10.1016/B978-0-12-407704-1.00005-1. Int Rev Cell Mol Biol. 2013. PMID: 23317820 Free PMC article. Review. - Epithelial Xbp1 is required for cellular proliferation and differentiation during mammary gland development.
Hasegawa D, Calvo V, Avivar-Valderas A, Lade A, Chou HI, Lee YA, Farias EF, Aguirre-Ghiso JA, Friedman SL. Hasegawa D, et al. Mol Cell Biol. 2015 May;35(9):1543-56. doi: 10.1128/MCB.00136-15. Epub 2015 Feb 23. Mol Cell Biol. 2015. PMID: 25713103 Free PMC article. - Platelet-activating factor acetyl hydrolase IB2 dysregulated cell proliferation in ovarian cancer.
He Y, He Z, Zhang X, Liu S. He Y, et al. Cancer Cell Int. 2021 Dec 20;21(1):697. doi: 10.1186/s12935-021-02406-9. Cancer Cell Int. 2021. PMID: 34930255 Free PMC article. - Enzymatic Characterization of ER Stress-Dependent Kinase, PERK, and Development of a High-Throughput Assay for Identification of PERK Inhibitors.
Pytel D, Seyb K, Liu M, Ray SS, Concannon J, Huang M, Cuny GD, Diehl JA, Glicksman MA. Pytel D, et al. J Biomol Screen. 2014 Aug;19(7):1024-34. doi: 10.1177/1087057114525853. Epub 2014 Mar 5. J Biomol Screen. 2014. PMID: 24598103 Free PMC article. - ER stress activates TAp73α to promote colon cancer cell apoptosis via the PERK-ATF4 pathway.
Sun S, Yi Y, Xiao ZJ, Chen H. Sun S, et al. J Cancer. 2023 Jul 3;14(11):1946-1955. doi: 10.7150/jca.84170. eCollection 2023. J Cancer. 2023. PMID: 37497416 Free PMC article.
References
- Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, Cook JL. Nrf2, a Cap‘n’Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem. 1999;274:26071–8. - PubMed
- Bartkova JHZ, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Orntoft T, Lukas J, Bartek J. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–70. - PubMed
- Bassing CH, Alt FW. H2AX may function as an anchor to hold broken chromosomal DNA ends in close proximity. Cell Cycle. 2004;3:149–53. - PubMed
- Behrend L, Henderson G, Zwacka RM. Reactive oxygen species in oncogenic transformation. Biochem Soc Trans. 2003;31:1441–4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 CA104838/CA/NCI NIH HHS/United States
- P01 CA104838-050003/CA/NCI NIH HHS/United States
- R01 DK088140/DK/NIDDK NIH HHS/United States
- F32CA1238252/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials