Computational analysis of unassigned high-quality MS/MS spectra in proteomic data sets - PubMed (original) (raw)
Computational analysis of unassigned high-quality MS/MS spectra in proteomic data sets
Kang Ning et al. Proteomics. 2010 Jul.
Abstract
In a typical shotgun proteomics experiment, a significant number of high-quality MS/MS spectra remain "unassigned." The main focus of this work is to improve our understanding of various sources of unassigned high-quality spectra. To achieve this, we designed an iterative computational approach for more efficient interrogation of MS/MS data. The method involves multiple stages of database searching with different search parameters, spectral library searching, blind searching for modified peptides, and genomic database searching. The method is applied to a large publicly available shotgun proteomic data set.
Figures
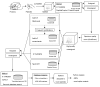
Figure 1. Overview of the iterative peptide identification strategy
Proteins are digested into peptides, and peptides are sequenced using MS/MS. Acquired spectra are analyzed using conventional database searching. Peptide identifications are processed using PeptideProphet and ProteinProphet. A spectral quality assessment tool is used to select unassigned high quality spectra. These spectra are reanalyzed using X! TANDEM and InsPecT (normal and blind mode) against the subset protein database, and using SpectraST spectral library search tool. The remaining unassigned spectra are searched against the translated genomic database to identify novel peptides and peptide polymorphisms.
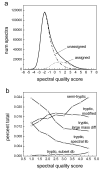
Figure 2. Prevalence and categories of unassigned high quality spectra
(a) The distribution of spectral quality scores plotted for all spectra (solid line), and separately for unassigned (dash dot line) and assigned (short dash) spectra after the initial database search. (b) The ratio of spectra assigned to peptides of different types (“percent total” refers to the proportion of spectra assigned to peptides of different type among the total number of initially unassigned spectra) during reanalysis, plotted as a function of the spectral quality score. The category ‘tryptic, subset db’ refers to spectra corresponding to unmodified tryptic peptides that were identified due to reduced search space. The category ‘tryptic, spectral lib’ refers to spectra corresponding to unmodified tryptic peptides identified using spectral library searching, and includes some spectra that were also identified by other methods. WCL fraction data.
Figure 3. Additional analysis of peptide categories
(a) The ratio of proteins (among proteins of similar abundance as measured using spectral counts) containing at least one modified peptide of a particular type (WCL fraction data). Shown are methionine oxidation (+16), N-terminal acetylation/carbamylation (+42), and pyroglutamic acid formation from N terminal glutamic acid (−17.0) (b) Most frequent modifications and their normalized frequencies in WCL, plasma membrane (PM), and raft fractions. (c) Novel peptides (according to NCBI NR database) identified by the genomic database search and categorized by edit distance (WCL, plasma membrane, raft fractions).
Similar articles
- Unassigned MS/MS Spectra: Who Am I?
Pathan M, Samuel M, Keerthikumar S, Mathivanan S. Pathan M, et al. Methods Mol Biol. 2017;1549:67-74. doi: 10.1007/978-1-4939-6740-7_6. Methods Mol Biol. 2017. PMID: 27975284 - Combination of Multiple Spectral Libraries Improves the Current Search Methods Used to Identify Missing Proteins in the Chromosome-Centric Human Proteome Project.
Cho JY, Lee HJ, Jeong SK, Kim KY, Kwon KH, Yoo JS, Omenn GS, Baker MS, Hancock WS, Paik YK. Cho JY, et al. J Proteome Res. 2015 Dec 4;14(12):4959-66. doi: 10.1021/acs.jproteome.5b00578. Epub 2015 Sep 14. J Proteome Res. 2015. PMID: 26330117 - A semi-empirical approach for predicting unobserved peptide MS/MS spectra from spectral libraries.
Hu Y, Li Y, Lam H. Hu Y, et al. Proteomics. 2011 Dec;11(24):4702-11. doi: 10.1002/pmic.201100316. Epub 2011 Nov 23. Proteomics. 2011. PMID: 22038894 - Current algorithmic solutions for peptide-based proteomics data generation and identification.
Hoopmann MR, Moritz RL. Hoopmann MR, et al. Curr Opin Biotechnol. 2013 Feb;24(1):31-8. doi: 10.1016/j.copbio.2012.10.013. Epub 2012 Nov 8. Curr Opin Biotechnol. 2013. PMID: 23142544 Free PMC article. Review. - Building and searching tandem mass (MS/MS) spectral libraries for peptide identification in proteomics.
Lam H, Aebersold R. Lam H, et al. Methods. 2011 Aug;54(4):424-31. doi: 10.1016/j.ymeth.2011.01.007. Epub 2011 Jan 28. Methods. 2011. PMID: 21277371 Review.
Cited by
- MSblender: A probabilistic approach for integrating peptide identifications from multiple database search engines.
Kwon T, Choi H, Vogel C, Nesvizhskii AI, Marcotte EM. Kwon T, et al. J Proteome Res. 2011 Jul 1;10(7):2949-58. doi: 10.1021/pr2002116. Epub 2011 Apr 29. J Proteome Res. 2011. PMID: 21488652 Free PMC article. - Common errors in mass spectrometry-based analysis of post-translational modifications.
Kim MS, Zhong J, Pandey A. Kim MS, et al. Proteomics. 2016 Mar;16(5):700-14. doi: 10.1002/pmic.201500355. Proteomics. 2016. PMID: 26667783 Free PMC article. Review. - Combining results of multiple search engines in proteomics.
Shteynberg D, Nesvizhskii AI, Moritz RL, Deutsch EW. Shteynberg D, et al. Mol Cell Proteomics. 2013 Sep;12(9):2383-93. doi: 10.1074/mcp.R113.027797. Epub 2013 May 29. Mol Cell Proteomics. 2013. PMID: 23720762 Free PMC article. Review. - ScanRanker: Quality assessment of tandem mass spectra via sequence tagging.
Ma ZQ, Chambers MC, Ham AJ, Cheek KL, Whitwell CW, Aerni HR, Schilling B, Miller AW, Caprioli RM, Tabb DL. Ma ZQ, et al. J Proteome Res. 2011 Jul 1;10(7):2896-904. doi: 10.1021/pr200118r. Epub 2011 Apr 26. J Proteome Res. 2011. PMID: 21520941 Free PMC article.
References
- Nesvizhskii AI, Vitek O, Aebersold R. Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat Methods. 2007;4:787–797. - PubMed
- Hernandez P, Muller M, Appel RD. Automated protein identification by tandem mass spectrometry: Issues and strategies. Mass Spectrometry Reviews. 2006;25:235–254. - PubMed
- Nesvizhskii AI, Roos FF, Grossmann J, Vogelzang M, Eddes JS, Gruissem W, Baginsky S, Aebersold R. Dynamic spectrum quality assessment and iterative computational analysis of shotgun proteomic data: toward more efficient identification of post-translational modifications, sequence polymorphisms, and novel peptides. Mol Cell Proteomics. 2006;5:652–670. - PubMed
- Flikka K, Martens L, Vandekerckhoe J, Gevaert K, Eidhammer I. Improving the reliability and throughput of mass spectrometry-based proteomics by spectrum quality filtering. Proteomics. 2006;6:2086–2094. - PubMed
- Moore RE, Young MK, Lee TD. Method for screening peptide fragment ion mass spectra prior to database searching. Journal of the American Society for Mass Spectrometry. 2000;11:422–426. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources