SF1 and SF2 helicases: family matters - PubMed (original) (raw)
Review
SF1 and SF2 helicases: family matters
Margaret E Fairman-Williams et al. Curr Opin Struct Biol. 2010 Jun.
Abstract
Helicases of the superfamily (SF) 1 and 2 are involved in virtually all aspects of RNA and DNA metabolism. SF1 and SF2 helicases share a catalytic core with high structural similarity, but different enzymes even within each SF perform a wide spectrum of distinct functions on diverse substrates. To rationalize similarities and differences between these helicases, we outline a classification based on protein families that are characterized by typical sequence, structural, and mechanistic features. This classification complements and extends existing SF1 and SF2 helicase categorizations and highlights major structural and functional themes for these proteins. We discuss recent data in the context of this unifying view of SF1 and SF2 helicases.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures
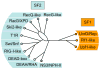
Figure 1. The Families of the SF1 and SF2 helicases
Schematic, unrooted cladogram showing the three identified families of the SF1 (right), and the 9 families and one group of the SF2 (left). A comprehensive list of proteins in each family in human, S_.cerevisiae_, and E.coli is given in the Supplementary Tables ST1 and ST2. Families were identified by a combination of phylogenetic analysis of the alignment of all SF1 and SF2 proteins from human, S.cerevisiae, E.coli, and selected viruses (Suppl. Fig. S1), and scoring for presence or absence of distinct sequence features and characteristic domain organization (for more detail, see Suppl. Figs. S1–3) and corresponding captions). Assignment of proteins to a family was highly robust. The phylogenetic relationships between the families were more ambiguous and tree topologies varied for some branches according to method and parameters used for generating the trees (Suppl. Fig. S2). The cladogram shown represents an average of multiple trees generated by different means (Suppl. Fig. S2). Branch lengths are not to scale. The oval indicates significant uncertainty in the tree topology in this region. Families were named according to names in use, or according to prominent members. Families named after a single protein were termed – like (i.e., Ski2-like). Non-standard abbreviations are as follows: T1R – type 1 restriction enzymes, RHA – RNA helicase A.
Figure 2. Sequence and structural organization of the helicase core of SF1 and SF2 proteins
(A) Sequence organization of the helicase core in SF1 and SF2. Characteristic sequence motifs are colored according to their predominant biochemical function: red, ATP binding and hydrolysis; yellow, coordination between nucleic acid and NTP binding sites; blue, nucleic acid binding. Green circled asterisks mark insertions of additional domains. The lengths of the blocks and the distance between the conserved domains is not to scale. Characteristic motifs were identified from the alignment of all SF1 and SF2 proteins from human, S.cerevisiae, E.coli and selected viruses (Suppl. Fig. S1). Considering numbering schemes already in use [7], motifs were numbered consecutively. Motifs in SF1 and SF2 proteins are located at identical positions in the RecA-fold (panel B, below). Motif IIIa in SF1 has been occasionally marked as motif IV. The Q-motif is equivalent to motif 0 in RecQ proteins. Motif IVa in SF2 proteins is frequently marked QxxR, motif Ic often TPGR. The asterisk on motif Ib indicates that in some proteins this motif is replaced by an additional domain. (B) Sequence conservation within the characteristic helicase motifs. The height of the amino acids reflects the level of conservation at a given position, tall letters indicate higher conservation. The universally conserved E in motif II corresponds to 4 bits. Coloring marks the chemical properties of a given amino acid position: green - polar, blue - basic, red – acidic, and black - hydrophobic. Sequence logos were created from the alignment of SF1 and SF2 proteins (Suppl. Fig. S1) according to reference [91]. Circles under the letters are for visual guidance. For sequence logos of the characteristic motifs of the individual families see Suppl. Fig. S3 (C) Position of the characteristic motifs in the RecA-like folds of the helicase core domains. The β-strands are indicated by arrows, α-helices by cylinders. The β-strands of the first RecA-like domain are numbered according to their position in the primary structure. The position of the characteristic motifs is indicated by numbered circles, colored as in panel A. The position of inserted domains are marked by green circled asterisks, as in panel A. Blue coloring of the rightmost β-strand and α-helix in the SF2 representation indicates the absence of this part in several SF2 protein families. (D) Position of the characteristic motifs in three-dimensional structures of SF1 and SF2 proteins. Structures of the SF1 helicase UvrD (left, UvrD/Rep family) and the SF2 helicase Vasa (right, DEAD-box family) [40,92]. The bound ATP analog is colored margenta, the nucleic acid is colored wheat. Conserved sequence motifs are colored as in panel (A), inserted domains 1B and 2B of UvrD are light-pink and light-green.
Figure 3. Domain organization of SF2 and SF1 helicase families and groups
Domains are not to scale. (a) C- and N-termini of DEAD-box proteins include RRMs, Zn-fingers, tudor domains and others [26]. (b) The family-typical domain inserted between the helicase domains is shown in grey. RIG-I-like proteins vary in their terminal domains [70]. Prominent RIG-I-like proteins are shown, Mph1p/FancM-related proteins are not shown. (CARD – caspase recruitment domain, RD – regulatory domain, a Zn-binding domain [67], PAZ – PIWI, Argonaute, Zwille, dsRBD – double strand RNA binding domain) (c) Domain organization of bacterial RapA [93]. Inserted domain between the helicase core domains is family-typical (Suppl. Fig. 1) (NTD – N-terminal domain, CTD – C terminal domain). (d) Domain organization of EcoR124 [80]. (e) RecQ-like proteins feature multiple C and N-terminal domains including exonuclease domains. Shown are the most conserved features of the RecQ-like family [94]. (Zn – Zn finger domain, WH – winged helix domain, HRDC – Helicase and RNAseD-like C terminal domain, RQC – RecQ C-terminal domain) (f) Rad3/XPD proteins also feature diverse C and N-termini. The domains inserted in helicase domain 1 are family-typical (FeS – iron-sulfur cluster, Arch – Arch domain, a structural domain, see Fig. 4) (g) Domain organization in RecG-like protein is largely conserved, but PriA has a non-typical Zn finger inserted in the helicase domain 2 [66] (TRG – translocation by RecG) (h) Domain organization of Hel 308 [52]. The organization of the C-terminal domains is conserved in Brr2 [95,96]. (WH – winged helix, H1 – helical 1, H2 – helical 2, FN3 – fibronectin 3) (i) DEAH/RHA proteins have varying N-terminal domains, but show a very high degree of conservation in their C-termini, especially among the spliceosomal DEAH proteins. It is possible that DEAH proteins and perhaps even most DEAH/RHA proteins show a conserved domain organization of their C-termini. Shown is the domain organization of Prp43 [53]. The domain organization of the C-terminus, with the exception of the OB-fold domain, resembles that of Ski2-like proteins [52,95,96]. (WH* - degenerated winged helix, Ratchet corresponds to H1 and H1 in the Ski2-like proteins) (k) NS3/NPH-II proteins have pronounced C- and N-terminal domains, but with the exception of the shown helical C-terminus of NS3 proteins from flaviviridae [51], no further information about these domains is available. (l) Upf1-like proteins feature variable termini. The location of domains 1B and 1C is conserved in the family. Shown is the domain organization of Upf1. (m) Pif1-like proteins feature variable termini, but location of domains 1B is conserved in the family. Domain 2B varies in size from about a dozen residues (RecD1) to more than 100 amino acids (Pif1p). (n) UvrD/Rep proteins also have variable termini. The location of domains 1B and 2B is largely conserved, but domain 2B is absent in LBA1 and HelD. LBA1 also features 3 ankyrin repeats inserted in the helicase domain 1 before domain 1B.
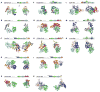
Figure 4. Domain architecture of SF2 and SF1 helicase families and groups
A representative structure of one protein from each family is shown, as indicated. The schematic cartoons show the domain organization of the displayed structure. In all structures, the helicase domain 1 (H1) is green, helicase domain 2 (H2) cyan, and nucleic acid, where present, beige. Terminal and inserted accessory domains are colored according to their respective folds, as indicated. All structures are oriented in a similar fashion, right panels show the structures rotated by roughly 90°, as indicated in panel (A). (Abbreviations: αH - helical domain, L24 fold – fold resembling the ribosomal protein L24, Zn – Zinc binding domain, WH – winged helix domain, FeS – iron-sulphur cluster, OB – OB fold, TRG – translocation by RecG) 1B,C and 2B mark the inserts in SF1 proteins. The following structures are shown: (A) Mss116p (S.cerevisiae) [44], (B) Hef (Pyrococcus furiosis) [63], (C) RapA (E.coli) [93], (D) EcoR124I (E.coli) [80], (E) RecQ1 (H.sapiens) [55], (F) XPD (Sulfolobus acidocaldarius) [58], (G) Hel308 (Archaeoglobus fulgidus) [52], (H) Prp43p (S.cerevisiae) [53], (I) HCV NS3 (Hepatitis C virus) [97], (K) RecG (Thermotoga maritima) [98], (L) Upf1 (H.sapiens) [99], (M) RecD2 (D.radiourans) [41], (N) UvrD (E.coli) [40].
Similar articles
- Genome-wide identification of SF1 and SF2 helicases from archaea.
Chamieh H, Ibrahim H, Kozah J. Chamieh H, et al. Gene. 2016 Jan 15;576(1 Pt 2):214-28. doi: 10.1016/j.gene.2015.10.007. Epub 2015 Oct 8. Gene. 2016. PMID: 26456193 - Comparisons between the structures of HCV and Rep helicases reveal structural similarities between SF1 and SF2 super-families of helicases.
Korolev S, Yao N, Lohman TM, Weber PC, Waksman G. Korolev S, et al. Protein Sci. 1998 Mar;7(3):605-10. doi: 10.1002/pro.5560070309. Protein Sci. 1998. PMID: 9541392 Free PMC article. - Binding and unwinding: SF3 viral helicases.
Hickman AB, Dyda F. Hickman AB, et al. Curr Opin Struct Biol. 2005 Feb;15(1):77-85. doi: 10.1016/j.sbi.2004.12.001. Curr Opin Struct Biol. 2005. PMID: 15718137 Review. - An ancient anion-binding structural module in RNA and DNA helicases.
Milner-White EJ, Pietras Z, Luisi BF. Milner-White EJ, et al. Proteins. 2010 Jun;78(8):1900-8. doi: 10.1002/prot.22704. Proteins. 2010. PMID: 20310069 Free PMC article. - Superfamily 2 helicases.
Byrd AK, Raney KD. Byrd AK, et al. Front Biosci (Landmark Ed). 2012 Jun 1;17(6):2070-88. doi: 10.2741/4038. Front Biosci (Landmark Ed). 2012. PMID: 22652765 Free PMC article. Review.
Cited by
- Molecular mechanics of RNA translocases.
Ding SC, Pyle AM. Ding SC, et al. Methods Enzymol. 2012;511:131-47. doi: 10.1016/B978-0-12-396546-2.00006-1. Methods Enzymol. 2012. PMID: 22713318 Free PMC article. - Steady-state NTPase activity of Dengue virus NS3: number of catalytic sites, nucleotide specificity and activation by ssRNA.
Incicco JJ, Gebhard LG, González-Lebrero RM, Gamarnik AV, Kaufman SB. Incicco JJ, et al. PLoS One. 2013;8(3):e58508. doi: 10.1371/journal.pone.0058508. Epub 2013 Mar 19. PLoS One. 2013. PMID: 23526990 Free PMC article. - Evaluation of the potency of FDA-approved drugs on wild type and mutant SARS-CoV-2 helicase (Nsp13).
Ugurel OM, Mutlu O, Sariyer E, Kocer S, Ugurel E, Inci TG, Ata O, Turgut-Balik D. Ugurel OM, et al. Int J Biol Macromol. 2020 Nov 15;163:1687-1696. doi: 10.1016/j.ijbiomac.2020.09.138. Epub 2020 Sep 24. Int J Biol Macromol. 2020. PMID: 32980406 Free PMC article. - DDX3 DEAD-box RNA helicase plays a central role in mitochondrial protein quality control in Leishmania.
Padmanabhan PK, Zghidi-Abouzid O, Samant M, Dumas C, Aguiar BG, Estaquier J, Papadopoulou B. Padmanabhan PK, et al. Cell Death Dis. 2016 Oct 13;7(10):e2406. doi: 10.1038/cddis.2016.315. Cell Death Dis. 2016. PMID: 27735940 Free PMC article. - Unwinding the mechanisms of a DEAD-box RNA helicase in cancer.
Russell R. Russell R. J Mol Biol. 2015 May 8;427(9):1797-800. doi: 10.1016/j.jmb.2015.03.009. Epub 2015 Mar 30. J Mol Biol. 2015. PMID: 25836982 Free PMC article. No abstract available.
References
- Abdel-Monem M, Hoffman-Berling H. Enzymic Unwinding of DNA 2. Chain separtion by an ATP-dependent DNA unwinding enyzme. Eur J Biochem. 1976;65:441–449. - PubMed
- Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Cell Biol. 2008;9:391–401. - PubMed
- Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Ann Rev Biochem. 2007;76:23–50. - PubMed
- Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Ann Rev Biophys. 2008;37:317–336. - PubMed
- Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317:41–72. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases