Dali server: conservation mapping in 3D - PubMed (original) (raw)
. 2010 Jul;38(Web Server issue):W545-9.
doi: 10.1093/nar/gkq366. Epub 2010 May 10.
Affiliations
- PMID: 20457744
- PMCID: PMC2896194
- DOI: 10.1093/nar/gkq366
Dali server: conservation mapping in 3D
Liisa Holm et al. Nucleic Acids Res. 2010 Jul.
Abstract
Our web site (http://ekhidna.biocenter.helsinki.fi/dali\_server) runs the Dali program for protein structure comparison. The web site consists of three parts: (i) the Dali server compares newly solved structures against structures in the Protein Data Bank (PDB), (ii) the Dali database allows browsing precomputed structural neighbourhoods and (iii) the pairwise comparison generates suboptimal alignments for a pair of structures. Each part has its own query form and a common format for the results page. The inputs are either PDB identifiers or novel structures uploaded by the user. The results pages are hyperlinked to aid interactive analysis. The web interface is simple and easy to use. The key purpose of interactive analysis is to check whether conserved residues line up in multiple structural alignments and how conserved residues and ligands cluster together in multiple structure superimpositions. In favourable cases, protein structure comparison can lead to evolutionary discoveries not detected by sequence analysis.
Figures
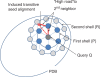
Figure 1.
The walking strategy focuses the search for structural neighbours to the neighbourhood of similar structures found so far. A sparse network of alignments within PDB is stored in an internal database. Once query Q is aligned to some PDB structure(s), a complete set of similar structures can be collected by walking, provided that the network is connected and similarity is approximately transitive.
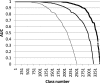
Figure 2.
Benchmarking the walking strategy. The plots show the distribution of AUC values for single-domain queries in scop 1.75. AUC, where coverage is TP/T and reliability is TP/P with T the number of members of a scop class, P number of matches above a given _Z_-score, and TP the number of members of a scop class above a given _Z_-score. Evaluated classes are scop folds (thin line), scop superfamilies (medium thick line) and scop families (thick line). Classes with fewer than 10 or more than 500 members are excluded. Query structures consisted of single domains from scop classes a-d, filtered at 90% sequence identity. Matches to all PDB structures which contained an instance of a domain in scop classes a-d were included in the evaluation.
Figure 3.
Using 3kk7A as query, the Dali server produces several views of the result, including: match list (top left), sequence view (top right), secondary structure view (bottom right), mapping of structural conservation (bottom left) and mapping of sequence conservation (middle bottom). The match list is sorted by _Z_-scores. The sequence view uses colour for amino acids which have a frequency above 0.5 in a column. The secondary structure view displays the alignment of helices (H), strands (E) and loops (L) as defined by DSSP. Sequence and structural conservation is computed within the selected subset of matches; red positions are maximally conserved and blue positions are not conserved.
Similar articles
- Dali server update.
Holm L, Laakso LM. Holm L, et al. Nucleic Acids Res. 2016 Jul 8;44(W1):W351-5. doi: 10.1093/nar/gkw357. Epub 2016 Apr 29. Nucleic Acids Res. 2016. PMID: 27131377 Free PMC article. - Using Dali for Protein Structure Comparison.
Holm L. Holm L. Methods Mol Biol. 2020;2112:29-42. doi: 10.1007/978-1-0716-0270-6_3. Methods Mol Biol. 2020. PMID: 32006276 - Searching protein structure databases with DaliLite v.3.
Holm L, Kääriäinen S, Rosenström P, Schenkel A. Holm L, et al. Bioinformatics. 2008 Dec 1;24(23):2780-1. doi: 10.1093/bioinformatics/btn507. Epub 2008 Sep 25. Bioinformatics. 2008. PMID: 18818215 Free PMC article. - QSalignWeb: A Server to Predict and Analyze Protein Quaternary Structure.
Dey S, Prilusky J, Levy ED. Dey S, et al. Front Mol Biosci. 2022 Jan 5;8:787510. doi: 10.3389/fmolb.2021.787510. eCollection 2021. Front Mol Biosci. 2022. PMID: 35071324 Free PMC article. Review. - Comparison of proteins based on segments structural similarity.
Plewczynski D, Pas J, Von Grotthuss M, Rychlewski L. Plewczynski D, et al. Acta Biochim Pol. 2004;51(1):161-72. Acta Biochim Pol. 2004. PMID: 15094837 Review.
Cited by
- Structural insight into the distinct regulatory mechanism of the HEPN-MNT toxin-antitoxin system in Legionella pneumophila.
Jin C, Jeon CH, Kim HW, Kang JM, Choi Y, Kang SM, Lee HH, Kim DH, Han BW, Lee BJ. Jin C, et al. Nat Commun. 2024 Nov 24;15(1):10188. doi: 10.1038/s41467-024-54551-0. Nat Commun. 2024. PMID: 39582057 Free PMC article. - Discovery of Five Classes of Bacterial Defensins: Ancestral Precursors of Defensins from Eukarya?
Cardoso MH, de Lima LR, Pires AS, Maximiano MR, Harvey PJ, Freitas CG, Costa RA, Fensterseifer ICM, Rigueiras PO, Migliolo L, Porto WF, Craik DJ, Franco OL. Cardoso MH, et al. ACS Omega. 2024 Oct 11;9(45):45297-45308. doi: 10.1021/acsomega.4c06956. eCollection 2024 Nov 12. ACS Omega. 2024. PMID: 39554447 Free PMC article. - A novel esterase regulates Klebsiella pneumoniae hypermucoviscosity and virulence.
Wang L, Wang Z, Zhang H, Jin Q, Fan S, Liu Y, Huang X, Guo J, Cai C, Zhang JR, Wu H. Wang L, et al. PLoS Pathog. 2024 Oct 31;20(10):e1012675. doi: 10.1371/journal.ppat.1012675. eCollection 2024 Oct. PLoS Pathog. 2024. PMID: 39480904 Free PMC article. - Unique Fn3-like biosensor in σI/anti-σI factors for regulatory expression of major cellulosomal scaffoldins in Pseudobacteroides cellulosolvens.
Dong S, Chen C, Li J, Liu YJ, Bayer EA, Lamed R, Mizrahi I, Cui Q, Feng Y. Dong S, et al. Protein Sci. 2024 Nov;33(11):e5193. doi: 10.1002/pro.5193. Protein Sci. 2024. PMID: 39470320
References
- Hasegawa H, Holm L. Advances and pitfalls of protein structural alignment. Curr. Opin. Struct. Biol. 2009;19:381–389. - PubMed
- Kawabata T, Nishikawa K. Protein tertiary structure comparison using the Markov transition model of evolution. Proteins. 2000;41:108–122. - PubMed
- Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Cryst. 2004;D60:2256–2268. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources