MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data - PubMed (original) (raw)
. 2010 Jul;38(Web Server issue):W71-7.
doi: 10.1093/nar/gkq329. Epub 2010 May 10.
Affiliations
- PMID: 20457745
- PMCID: PMC2896187
- DOI: 10.1093/nar/gkq329
MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data
Jianguo Xia et al. Nucleic Acids Res. 2010 Jul.
Abstract
Gene set enrichment analysis (GSEA) is a widely used technique in transcriptomic data analysis that uses a database of predefined gene sets to rank lists of genes from microarray studies to identify significant and coordinated changes in gene expression data. While GSEA has been playing a significant role in understanding transcriptomic data, no similar tools are currently available for understanding metabolomic data. Here, we introduce a web-based server, called Metabolite Set Enrichment Analysis (MSEA), to help researchers identify and interpret patterns of human or mammalian metabolite concentration changes in a biologically meaningful context. Key to the development of MSEA has been the creation of a library of approximately 1000 predefined metabolite sets covering various metabolic pathways, disease states, biofluids, and tissue locations. MSEA also supports user-defined or custom metabolite sets for more specialized analysis. MSEA offers three different enrichment analyses for metabolomic studies including overrepresentation analysis (ORA), single sample profiling (SSP) and quantitative enrichment analysis (QEA). ORA requires only a list of compound names, while SSP and QEA require both compound names and compound concentrations. MSEA generates easily understood graphs or tables embedded with hyperlinks to relevant pathway images and disease descriptors. For non-mammalian or more specialized metabolomic studies, MSEA allows users to provide their own metabolite sets for enrichment analysis. The MSEA server also supports conversion between metabolite common names, synonyms, and major database identifiers. MSEA has the potential to help users identify obvious as well as 'subtle but coordinated' changes among a group of related metabolites that may go undetected with conventional approaches. MSEA is freely available at http://www.msea.ca.
Figures
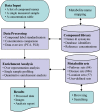
Figure 1.
MSEA workflow. MSEA consists of four steps—data input, data processing, data analysis, and data download. Different analysis procedures are performed for different input types. MSEA allows users to directly browse and search its metabolite set libraries as well as to perform metabolite name mapping between different names and database ID.
Figure 2.
Enrichment analysis and visualization. Results from MSEA’s enrichment analysis are presented both in tables as well as through graphical summaries. (A) The comparison between the measured concentrations and reference concentrations using the SSP module. The top part of (B) shows a graphical summary of the concentration comparison for a single compound when users click an image icon in Figure 2A. The bottom part of Figure 2B shows all the corresponding publications that reported these concentrations. (C) The results generated by the QEA module. The top part of (D) is a metabolite-set plot indicating the influence of an individual compound on each of the selected metabolite sets. The bottom part of Figure 2D shows all its constituent metabolites with matched ones highlighted in red.
Similar articles
- Metabolomic data processing, analysis, and interpretation using MetaboAnalyst.
Xia J, Wishart DS. Xia J, et al. Curr Protoc Bioinformatics. 2011 Jun;Chapter 14:14.10.1-14.10.48. doi: 10.1002/0471250953.bi1410s34. Curr Protoc Bioinformatics. 2011. PMID: 21633943 - MetaboAnalyst: a web server for metabolomic data analysis and interpretation.
Xia J, Psychogios N, Young N, Wishart DS. Xia J, et al. Nucleic Acids Res. 2009 Jul;37(Web Server issue):W652-60. doi: 10.1093/nar/gkp356. Epub 2009 May 8. Nucleic Acids Res. 2009. PMID: 19429898 Free PMC article. - Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis.
Chong J, Wishart DS, Xia J. Chong J, et al. Curr Protoc Bioinformatics. 2019 Dec;68(1):e86. doi: 10.1002/cpbi.86. Curr Protoc Bioinformatics. 2019. PMID: 31756036 - Translational Metabolomics of Head Injury: Exploring Dysfunctional Cerebral Metabolism with Ex Vivo NMR Spectroscopy-Based Metabolite Quantification.
Wolahan SM, Hirt D, Glenn TC. Wolahan SM, et al. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. PMID: 26269925 Free Books & Documents. Review. - Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis.
Xia J, Wishart DS. Xia J, et al. Curr Protoc Bioinformatics. 2016 Sep 7;55:14.10.1-14.10.91. doi: 10.1002/cpbi.11. Curr Protoc Bioinformatics. 2016. PMID: 27603023 Review.
Cited by
- Patients With Common Variable Immunodeficiency (CVID) Show Higher Gut Bacterial Diversity and Levels of Low-Abundance Genes Than the Healthy Housemates.
Bosák J, Lexa M, Fiedorová K, Gadara DC, Micenková L, Spacil Z, Litzman J, Freiberger T, Šmajs D. Bosák J, et al. Front Immunol. 2021 May 14;12:671239. doi: 10.3389/fimmu.2021.671239. eCollection 2021. Front Immunol. 2021. PMID: 34054845 Free PMC article. - A comprehensive IDA and SWATH-DIA Lipidomics and Metabolomics dataset: SARS-CoV-2 case control study.
Tahir A, Draxler A, Stelzer T, Blaschke A, Laky B, Széll M, Binar J, Bartak V, Bragagna L, Maqboul L, Herzog T, Thell R, Wagner KH. Tahir A, et al. Sci Data. 2024 Sep 12;11(1):998. doi: 10.1038/s41597-024-03822-y. Sci Data. 2024. PMID: 39266559 Free PMC article. - Plasma metabolomic and lipidomic profiles accurately classify mothers of children with congenital heart disease: an observational study.
Mires S, Sommella E, Merciai F, Salviati E, Caponigro V, Basilicata MG, Marini F, Campiglia P, Baquedano M, Dong T, Skerritt C, Eastwood KA, Caputo M. Mires S, et al. Metabolomics. 2024 Jul 2;20(4):70. doi: 10.1007/s11306-024-02129-8. Metabolomics. 2024. PMID: 38955892 Free PMC article. - MarVis-Pathway: integrative and exploratory pathway analysis of non-targeted metabolomics data.
Kaever A, Landesfeind M, Feussner K, Mosblech A, Heilmann I, Morgenstern B, Feussner I, Meinicke P. Kaever A, et al. Metabolomics. 2015;11(3):764-777. doi: 10.1007/s11306-014-0734-y. Epub 2014 Oct 10. Metabolomics. 2015. PMID: 25972773 Free PMC article. - Heat-Killed Lactobacillus acidophilus Promotes Growth by Modulating the Gut Microbiota Composition and Fecal Metabolites of Piglets.
Miao H, Liang J, Lan G, Wu Q, Huang Z. Miao H, et al. Animals (Basel). 2024 Aug 30;14(17):2528. doi: 10.3390/ani14172528. Animals (Basel). 2024. PMID: 39272313 Free PMC article.
References
- Wishart DS. Quantitative metabolomics using NMR. Trends Anal. Chem. 2008;27:228–237.
- Hollywood K, Brison DR, Goodacre R. Metabolomics: current technologies and future trends. Proteomics. 2006;6:4716–4723. - PubMed
- Ewald JC, Heux S, Zamboni N. High-throughput quantitative metabolomics: workflow for cultivation, quenching, and analysis of yeast in a multiwell format. Anal. Chem. 2009;81:3623–3629. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources