Gene expression profile and genomic alterations in colonic tumours induced by 1,2-dimethylhydrazine (DMH) in rats - PubMed (original) (raw)
Gene expression profile and genomic alterations in colonic tumours induced by 1,2-dimethylhydrazine (DMH) in rats
Angelo Pietro Femia et al. BMC Cancer. 2010.
Abstract
Background: Azoxymethane (AOM) or 1,2-dimethylhydrazine (DMH)-induced colon carcinogenesis in rats shares many phenotypical similarities with human sporadic colon cancer and is a reliable model for identifying chemopreventive agents. Genetic mutations relevant to human colon cancer have been described in this model, but comprehensive gene expression and genomic analysis have not been reported so far. Therefore, we applied genome-wide technologies to study variations in gene expression and genomic alterations in DMH-induced colon cancer in F344 rats.
Methods: For gene expression analysis, 9 tumours (TUM) and their paired normal mucosa (NM) were hybridized on 4 x 44K Whole rat arrays (Agilent) and selected genes were validated by semi-quantitative RT-PCR. Functional analysis on microarray data was performed by GenMAPP/MappFinder analysis. Array-comparative genomic hybridization (a-CGH) was performed on 10 paired TUM-NM samples hybridized on Rat genome arrays 2 x 105K (Agilent) and the results were analyzed by CGH Analytics (Agilent).
Results: Microarray gene expression analysis showed that Defcr4, Igfbp5, Mmp7, Nos2, S100A8 and S100A9 were among the most up-regulated genes in tumours (Fold Change (FC) compared with NM: 183, 48, 39, 38, 36 and 32, respectively), while Slc26a3, Mptx, Retlna and Muc2 were strongly down-regulated (FC: -500; -376, -167, -79, respectively). Functional analysis showed that pathways controlling cell cycle, protein synthesis, matrix metalloproteinases, TNFalpha/NFkB, and inflammatory responses were up-regulated in tumours, while Krebs cycle, the electron transport chain, and fatty acid beta oxidation were down-regulated. a-CGH analysis showed that four TUM out of ten had one or two chromosomal aberrations. Importantly, one sample showed a deletion on chromosome 18 including Apc.
Conclusion: The results showed complex gene expression alterations in adenocarcinomas encompassing many altered pathways. While a-CGH analysis showed a low degree of genomic imbalance, it is interesting to note that one of the alterations concerned Apc, a key gene in colorectal carcinogenesis. The fact that many of the molecular alterations described in this study are documented in human colon tumours confirms the relevance of DMH-induced cancers as a powerful tool for the study of colon carcinogenesis and chemoprevention.
Figures
Figure 1
Immunohistochemistry for NFkB/p65 and histology in a DMH-induced cancer. Panel A: adenocarcinoma sample challenged with an antibody against NFkB/p65. Panel B: magnification of the inset in panel A; note that positive cells in the tumour show immunoreactivity in both the cytoplasm and the nucleus. Panel C: paired normal mucosa of the tumor challenged with the same antibody. Panel D: H&E stained section of the same tumour. Original magnification in panels A and C: ×400, panel B: ×1000, panel D: ×100.
Figure 2
Semi-quantitative RT-PCR analysis of ten selected genes. For each up-regulated (first five rows) or down-regulated (last five rows) genes, three representative gel images of paired normal mucosa (N) and tumour (T) were provided. For each RT-PCR experiment, expression of β-actin was used as an internal control. P values refer to the t-test for paired samples (n = 7) to compare tumours with normal mucosa.
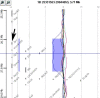
Figure 3
CGH analytics chromosome view of a region of chromosome 18 of all tumor samples analyzed. Colored curves represent Log2 ratio values for all nucleotide probes plotted as a function of their chromosomal position for each tumor sample. Thick straight vertical blue line and blue shaded area point out a deletion present in sample #9. The black arrow indicates the Apc gene position.
Similar articles
- Red wine polyphenols influence carcinogenesis, intestinal microflora, oxidative damage and gene expression profiles of colonic mucosa in F344 rats.
Dolara P, Luceri C, De Filippo C, Femia AP, Giovannelli L, Caderni G, Cecchini C, Silvi S, Orpianesi C, Cresci A. Dolara P, et al. Mutat Res. 2005 Dec 11;591(1-2):237-46. doi: 10.1016/j.mrfmmm.2005.04.022. Epub 2005 Nov 15. Mutat Res. 2005. PMID: 16293270 - Calcium prevents tumorigenesis in a mouse model of colorectal cancer.
Wang JL, Lin YW, Chen HM, Kong X, Xiong H, Shen N, Hong J, Fang JY. Wang JL, et al. PLoS One. 2011;6(8):e22566. doi: 10.1371/journal.pone.0022566. Epub 2011 Aug 17. PLoS One. 2011. PMID: 21857934 Free PMC article. - Morphological and molecular alterations in 1,2 dimethylhydrazine and azoxymethane induced colon carcinogenesis in rats.
Perše M, Cerar A. Perše M, et al. J Biomed Biotechnol. 2011;2011:473964. doi: 10.1155/2011/473964. Epub 2010 Dec 28. J Biomed Biotechnol. 2011. PMID: 21253581 Free PMC article. Review. - Three Pathways of Colonic Carcinogenesis in Rats.
Rubio CA. Rubio CA. Anticancer Res. 2017 Jan;37(1):15-20. doi: 10.21873/anticanres.11284. Anticancer Res. 2017. PMID: 28011469 Review.
Cited by
- The Antioxidant and Antitumor Efficiency of Litophyton sp. Extract in DMH-Induced Colon Cancer in Male Rats.
Ashry M, Askar H, Alian A, Zidan SAH, El-Sahra DG, Abdel-Wahhab KG, Lamlom SF, Abdelsalam NR, Abd El-Hack ME, Gomaa HF. Ashry M, et al. Life (Basel). 2022 Sep 21;12(10):1470. doi: 10.3390/life12101470. Life (Basel). 2022. PMID: 36294905 Free PMC article. - Evaluation of chemopreventive potential of Strobilanthes crispus against colon cancer formation in vitro and in vivo.
Al-Henhena N, Khalifa SA, Ying RP, Ismail S, Hamadi R, Shawter AN, Idris AM, Azizan A, Al-Wajeeh NS, Abdulla MA, El-Seedi HR. Al-Henhena N, et al. BMC Complement Altern Med. 2015 Nov 25;15(1):419. doi: 10.1186/s12906-015-0926-7. BMC Complement Altern Med. 2015. PMID: 26608653 Free PMC article. - Mucin depleted foci, colonic preneoplastic lesions lacking Muc2, show up-regulation of Tlr2 but not bacterial infiltration.
Femia AP, Swidsinski A, Dolara P, Salvadori M, Amedei A, Caderni G. Femia AP, et al. PLoS One. 2012;7(1):e29918. doi: 10.1371/journal.pone.0029918. Epub 2012 Jan 5. PLoS One. 2012. PMID: 22242189 Free PMC article. - Hepatoprotective effect of Matricaria chamomilla aqueous extract against 1,2-Dimethylhydrazine-induced carcinogenic hepatic damage in mice.
Shebbo S, El Joumaa M, Kawach R, Borjac J. Shebbo S, et al. Heliyon. 2020 Jun 1;6(6):e04082. doi: 10.1016/j.heliyon.2020.e04082. eCollection 2020 Jun. Heliyon. 2020. PMID: 32509999 Free PMC article.
References
- Cardoso J, Boer J, Morreau H, Fodde R. Expression and genomic profiling of colorectal cancer. Biochim Biophys Acta. 2007;1775:103–37. - PubMed
- Nambiar PR, Nakanishi M, Gupta R, Cheung E, Firouzi A, Ma XJ, Flynn C, Dong M, Guda K, Levine J, Raja R, Achenie L, Rosenberg DW. Genetic signatures of high- and low-risk aberrant crypt foci in a mouse model of sporadic colon cancer. Cancer Res. 2004;64:6394–401. doi: 10.1158/0008-5472.CAN-04-0933. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous