Molecular mechanism of MLL PHD3 and RNA recognition by the Cyp33 RRM domain - PubMed (original) (raw)
Molecular mechanism of MLL PHD3 and RNA recognition by the Cyp33 RRM domain
Robert A Hom et al. J Mol Biol. 2010.
Abstract
The nuclear protein cyclophilin 33 (Cyp33) is a peptidyl-prolyl cis-trans isomerase that catalyzes cis-trans isomerization of the peptide bond preceding a proline and promotes folding and conformational changes in folded and unfolded proteins. The N-terminal RNA-recognition motif (RRM) domain of Cyp33 has been found to associate with the third plant homeodomain (PHD3) finger of the mixed lineage leukemia (MLL) proto-oncoprotein and a poly(A) RNA sequence. Here, we report a 1.9 A resolution crystal structure of the RRM domain of Cyp33 and describe the molecular mechanism of PHD3 and RNA recognition. The Cyp33 RRM domain folds into a five-stranded antiparallel beta-sheet and two alpha-helices. The RRM domain, but not the catalytic module of Cyp33, binds strongly to PHD3, exhibiting a 2 muM affinity as measured by isothermal titration calorimetry. NMR chemical shift perturbation (CSP) analysis and dynamics data reveal that the beta strands and the beta2-beta3 loop of the RRM domain are involved in the interaction with PHD3. Mutations in the PHD3-binding site or deletions in the beta2-beta3 loop lead to a significantly reduced affinity or abrogation of the interaction. The RNA-binding pocket of the Cyp33 RRM domain, mapped on the basis of NMR CSP and mutagenesis, partially overlaps with the PHD3-binding site, and RNA association is abolished in the presence of MLL PHD3. Full-length Cyp33 acts as a negative regulator of MLL-induced transcription and reduces the expression levels of MLL target genes MEIS1 and HOXA9. Together, these in vitro and in vivo data provide insight into the multiple functions of Cyp33 RRM and suggest a Cyp33-dependent mechanism for regulating the transcriptional activity of MLL.
Copyright (c) 2010 Elsevier Ltd. All rights reserved.
Figures
Figure 1
The crystal structure of the RRM domain of Cyp33 determined at 1.85 Å resolution. (a) Architecture of Cyp33: the amino-terminal RRM domain and the catalytic CYP domain. (b) Ribbon diagram of the RRM structure.
Figure 2
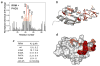
Binding of the RRM domain of Cyp33 to the PHD3 finger of MLL. (a) Schematic of MLL. The PHD3 finger is shown as a white oval. (b) Superimposed 1H,15N HSQC spectra of the 15N-labeled Cyp33 RRM domain, collected in the absence and presence of a two-fold excess of unlabeled MLL PHD3. (c) Superimposed 1H,15N HSQC spectra of the 15N-labeled MLL PHD3 finger, collected in the absence and presence of a two-fold excess of unlabeled Cyp33 RRM. (c) Representative ITC curves used to calculate the binding affinity for the interaction between the Cyp33 RRM domain and the MLL PHD3 finger.
Figure 3
The MLL PHD3-binding site of the Cyp33 RRM domain. (a) A histogram shows normalized 1H,15N chemical shift changes in backbone amides of 15N-labeled RRM upon addition of unlabeled PHD3 at a protein ratio of 1:1. (b, d) Residues that exhibit significant PHD3-induced resonance perturbations in (a) are mapped on the ribbon diagram (b) and the surface (d) of the Cyp33 RRM domain. Colored bars indicate significant change being greater than an average plus one standard deviation. (c) The binding affinities of wild type Cyp33 RRM and mutants for MLL PHD3, as measured by ITC. Interaction of the RRMΔ40–45 mutant was examined by NMR. Other mutants generated, including G48A, F54A, RRMΔ41–42, RRMΔ41–43 and RRMΔ40–43, precipitated during dialysis for ITC experiments.
Figure 4
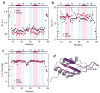
Dynamics of the Cyp33 RRM domain bound and unbound. (a–c) Relaxation parameters of the RRM domain in the absence (blue) and the presence (red) of a 1.5-fold excess of MLL PHD3. R1, R2 and NOE values were determined for backbone amide groups and are plotted for each residue of the Cyp33 RRM domain. The RRM secondary structure is shown above the graphs. (d) The most affected (due to the interaction with PHD3) residues of Cyp33 RRM are colored in shades of purple in the ribbon diagram of the RRM structure.
Figure 5
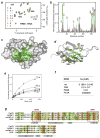
The RNA-binding site of the Cyp33 RRM domain. (a) Eight superimposed 1H,15N HSQC spectra of 0.1 mM Cyp33 RRM, collected as the AAUAAA RNA construct was gradually added. The spectra are color-coded according to the molar protein-RNA ratio (inset). (b) A histogram shows normalized 1H,15N chemical shift changes in backbone amides of 15N-labeled RRM (aa 1–90) upon addition of a six-fold excess of AAUAAA. (c, d) Residues that exhibit significant RNA-induced resonance perturbations in (b) are labeled and colored in shades of green on the ribbon diagram of the structure (c) and on the surface (d) of the Cyp33 RRM domain (aa 1–83). Colored bars indicate significant change being greater than an average plus one and a half standard deviation. (e) Representative binding curves used to determine the Kd values of the Cyp33 RRM-RNA interaction by NMR spectroscopy. (f) The RNA binding affinities of the wild type and mutant RRM domain measured by NMR. (g) Alignment of the RRM domain sequences of human proteins: absolutely, moderately and weakly conserved residues are colored brown, green and yellow respectively. The RNP2 and RNP1 motifs required for the interaction with RNA are outlined by red rectangles. The secondary structure of the Cyp33 RRM domain is shown below the sequences.
Figure 6
The MLL PHD3-binding site and the RNA-binding site of the Cyp33 RRM domain partially overlap. (a) The Cyp33 RRM domain is shown as a ribbon diagram. Residues of RRM that are perturbed by either PHD3, RNA or both ligands are colored red, green and yellow, respectively. The MLL PHD3-binding site and the RNA-binding site of the Cyp33 RRM domain are indicated by red and green circles, respectively. (b) Superimposed 1H,15N HSQC spectra of the Cyp33 RRM domain (0.1 mM) in the ligand-free form (black), after addition of 0.6 mM RNA (green), and after subsequent addition of 0.25 mM MLL PHD3 (red).
Figure 7
Cyp33 decreases MLL-dependent gene transcription. Gene expression levels were quantified in transfected 293T cells using quantitative real-time RT-PCR (a). (b) A model of the Cyp33-MLL association.
Similar articles
- Cyp33 binds AU-rich RNA motifs via an extended interface that competitively disrupts the gene repressive Cyp33-MLL1 interaction in vitro.
Lloyd NR, Wuttke DS. Lloyd NR, et al. PLoS One. 2021 Feb 19;16(2):e0237956. doi: 10.1371/journal.pone.0237956. eCollection 2021. PLoS One. 2021. PMID: 33606679 Free PMC article. - The PHD3 domain of MLL acts as a CYP33-regulated switch between MLL-mediated activation and repression.
Park S, Osmers U, Raman G, Schwantes RH, Diaz MO, Bushweller JH. Park S, et al. Biochemistry. 2010 Aug 10;49(31):6576-86. doi: 10.1021/bi1009387. Biochemistry. 2010. PMID: 20677832 Free PMC article. - Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression.
Wang Z, Song J, Milne TA, Wang GG, Li H, Allis CD, Patel DJ. Wang Z, et al. Cell. 2010 Jun 25;141(7):1183-94. doi: 10.1016/j.cell.2010.05.016. Epub 2010 Jun 10. Cell. 2010. PMID: 20541251 Free PMC article. - The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression.
Maris C, Dominguez C, Allain FH. Maris C, et al. FEBS J. 2005 May;272(9):2118-31. doi: 10.1111/j.1742-4658.2005.04653.x. FEBS J. 2005. PMID: 15853797 Review. - Integrated structural biology to unravel molecular mechanisms of protein-RNA recognition.
Schlundt A, Tants JN, Sattler M. Schlundt A, et al. Methods. 2017 Apr 15;118-119:119-136. doi: 10.1016/j.ymeth.2017.03.015. Epub 2017 Mar 16. Methods. 2017. PMID: 28315749 Review.
Cited by
- Multifunctional G-rich and RRM-containing domains of TbRGG2 perform separate yet essential functions in trypanosome RNA editing.
Foda BM, Downey KM, Fisk JC, Read LK. Foda BM, et al. Eukaryot Cell. 2012 Sep;11(9):1119-31. doi: 10.1128/EC.00175-12. Epub 2012 Jul 13. Eukaryot Cell. 2012. PMID: 22798390 Free PMC article. - Roles of Prolyl Isomerases in RNA-Mediated Gene Expression.
Thapar R. Thapar R. Biomolecules. 2015 May 18;5(2):974-99. doi: 10.3390/biom5020974. Biomolecules. 2015. PMID: 25992900 Free PMC article. Review. - A Structural Perspective on Readout of Epigenetic Histone and DNA Methylation Marks.
Patel DJ. Patel DJ. Cold Spring Harb Perspect Biol. 2016 Mar 1;8(3):a018754. doi: 10.1101/cshperspect.a018754. Cold Spring Harb Perspect Biol. 2016. PMID: 26931326 Free PMC article. Review. - Cyp33 binds AU-rich RNA motifs via an extended interface that competitively disrupts the gene repressive Cyp33-MLL1 interaction in vitro.
Lloyd NR, Wuttke DS. Lloyd NR, et al. PLoS One. 2021 Feb 19;16(2):e0237956. doi: 10.1371/journal.pone.0237956. eCollection 2021. PLoS One. 2021. PMID: 33606679 Free PMC article. - The PHD3 domain of MLL acts as a CYP33-regulated switch between MLL-mediated activation and repression.
Park S, Osmers U, Raman G, Schwantes RH, Diaz MO, Bushweller JH. Park S, et al. Biochemistry. 2010 Aug 10;49(31):6576-86. doi: 10.1021/bi1009387. Biochemistry. 2010. PMID: 20677832 Free PMC article.
References
- Mi H, Kops O, Zimmermann E, Jaschke A, Tropschug M. A nuclear RNA-binding cyclophilin in human T cells. FEBS Lett. 1996;398:201–5. - PubMed
- Wang XJ, Etzkorn FA. Peptidyl-prolyl isomerase inhibitors. Biopolymers. 2006;84:125–46. - PubMed
- Min L, Fulton DB, Andreotti AH. A case study of proline isomerization in cell signaling. Front Biosci. 2005;10:385–97. - PubMed
- Wang Y, Han R, Zhang W, Yuan Y, Zhang X, Long Y, Mi H. Human CyP33 binds specifically to mRNA and binding stimulates PPIase activity of hCyP33. FEBS Lett. 2008;582:835–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA116606/CA/NCI NIH HHS/United States
- CA55029/CA/NCI NIH HHS/United States
- R01 GM075827/GM/NIGMS NIH HHS/United States
- R01 CA055029/CA/NCI NIH HHS/United States
- CA116606/CA/NCI NIH HHS/United States
- U54 GM074961/GM/NIGMS NIH HHS/United States
- CA113472/CA/NCI NIH HHS/United States
- R01 GM071424/GM/NIGMS NIH HHS/United States
- GM075827/GM/NIGMS NIH HHS/United States
- GM074961/GM/NIGMS NIH HHS/United States
- GM071424/GM/NIGMS NIH HHS/United States
- R01 CA113472/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases