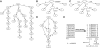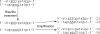A survey of sequence alignment algorithms for next-generation sequencing - PubMed (original) (raw)
Review
A survey of sequence alignment algorithms for next-generation sequencing
Heng Li et al. Brief Bioinform. 2010 Sep.
Abstract
Rapidly evolving sequencing technologies produce data on an unparalleled scale. A central challenge to the analysis of this data is sequence alignment, whereby sequence reads must be compared to a reference. A wide variety of alignment algorithms and software have been subsequently developed over the past two years. In this article, we will systematically review the current development of these algorithms and introduce their practical applications on different types of experimental data. We come to the conclusion that short-read alignment is no longer the bottleneck of data analyses. We also consider future development of alignment algorithms with respect to emerging long sequence reads and the prospect of cloud computing.
Figures
Figure 1:
Data structures based on a prefix trie. (A) Prefix trie of string AGGAGC where symbol ⁁ marks the start of the string. The two numbers in each node give the suffix array interval of the substring represented by the node, which is the string concatenation of edge symbols from the node to the root. (B) Compressed prefix trie by contracting nodes with in- and out-degree both being one. (C) Prefix tree by representing the substring on each edge as the interval on the original string. (D) Prefix directed word graph (prefix DAWG) created by collapsing nodes of the prefix trie with identical suffix array interval. (E) Constructing the suffix array and Burrows–Wheeler transform of AGGAGC. The dollar symbol marks the end of the string and is lexicographically smaller than all the other symbols. The suffix array interval of a substring W is the maximal interval in the suffix array with all suffixes in the interval having W as prefix. For example, the suffix array interval of AG is [1, 2]. The two suffixes in the interval are AGC$ and AGGAGC$, starting at position 3 and 0, respectively. They are the only suffixes that have AG as prefix.
Figure 2:
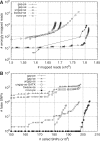
Alignment and SNP call accuracy under different configurations of BWA and Novoalign. (A) Number of misplaced reads as a function of the number of mapped reads under different mapping quality cut-off. Reads (108 bp) were simulated from human genome build36 assuming 0.085% substitution and 0.015% indel mutation rate, and 2% uniform sequencing error rate. (B) Number of wrong SNP calls as a function of the number of called SNP under different SNP quality cut-offs. Reads (108 bp) were simulated from chr6 of the human genome and mapped back to the whole genome. SNPs are called and filtered by SAMtools. In both figures, ‘novo-pe’ denotes novoalign alignment; the rest correspond to alignments under different configurations of BWA, where ‘gap-pe’ stands for the gapped paired-end (PE) alignment, ‘gap-se’ for gapped single-end (SE) alignment, ‘ungap-se’ for ungapped SE alignment, ‘bwasw-se’ for BWA-SW SE alignment, and ‘ungap-se-GATK’ for alignment cleaned by the GATK realigner.
Figure 3:
Alignment accuracy of simulated reads with and without base quality. Paired-end reads (51 bp) are simulated by MAQ from the human genome, assuming 0.085% substitution and 0.015% indel mutation rate. Base quality model is trained from run ERR000589 from the European short read archive. Base quality is not used in alignment for curves with labels ended with ‘-noQual’.
Figure 4:
Color-space encoding. (A) Color space encoding matrix. (B) Conversion between base and color sequence. (C) The color encoding of the reverse complement of the base sequence is the reverse of the color sequence. (D) A sequencing error leads to contiguous errors when the color sequence is converted to base sequence. (E) A mutation causes two contiguous color changes.
Figure 5:
Bisulfite sequencing. Cytosines with underlines are not methylated. Denaturation and bisulfite treatment will convert these cytosines to uracils. After amplification, four different sequences from the original double-strand DNA result.
Similar articles
- Long Read Alignment with Parallel MapReduce Cloud Platform.
Al-Absi AA, Kang DK. Al-Absi AA, et al. Biomed Res Int. 2015;2015:807407. doi: 10.1155/2015/807407. Epub 2015 Dec 29. Biomed Res Int. 2015. PMID: 26839887 Free PMC article. - Alignment of Next-Generation Sequencing Reads.
Reinert K, Langmead B, Weese D, Evers DJ. Reinert K, et al. Annu Rev Genomics Hum Genet. 2015;16:133-51. doi: 10.1146/annurev-genom-090413-025358. Epub 2015 May 4. Annu Rev Genomics Hum Genet. 2015. PMID: 25939052 Review. - Improved variant discovery through local re-alignment of short-read next-generation sequencing data using SRMA.
Homer N, Nelson SF. Homer N, et al. Genome Biol. 2010;11(10):R99. doi: 10.1186/gb-2010-11-10-r99. Epub 2010 Oct 8. Genome Biol. 2010. PMID: 20932289 Free PMC article. - RandAL: a randomized approach to aligning DNA sequences to reference genomes.
Vo NS, Tran Q, Niraula N, Phan V. Vo NS, et al. BMC Genomics. 2014;15 Suppl 5(Suppl 5):S2. doi: 10.1186/1471-2164-15-S5-S2. Epub 2014 Jul 14. BMC Genomics. 2014. PMID: 25081493 Free PMC article. - Sense from sequence reads: methods for alignment and assembly.
Flicek P, Birney E. Flicek P, et al. Nat Methods. 2009 Nov;6(11 Suppl):S6-S12. doi: 10.1038/nmeth.1376. Nat Methods. 2009. PMID: 19844229 Review.
Cited by
- When less is more: sketching with minimizers in genomics.
Ndiaye M, Prieto-Baños S, Fitzgerald LM, Yazdizadeh Kharrazi A, Oreshkov S, Dessimoz C, Sedlazeck FJ, Glover N, Majidian S. Ndiaye M, et al. Genome Biol. 2024 Oct 14;25(1):270. doi: 10.1186/s13059-024-03414-4. Genome Biol. 2024. PMID: 39402664 Free PMC article. Review. - TrialView: An AI-powered Visual Analytics System for Temporal Event Data in Clinical Trials.
Li Z, Liu X, Cheng Z, Chen Y, Tu W, Su J. Li Z, et al. Proc Annu Hawaii Int Conf Syst Sci. 2024;2024:1169-1178. Epub 2024 Jan 3. Proc Annu Hawaii Int Conf Syst Sci. 2024. PMID: 38681743 Free PMC article. - CGRWDL: alignment-free phylogeny reconstruction method for viruses based on chaos game representation weighted by dynamical language model.
Wang T, Yu ZG, Li J. Wang T, et al. Front Microbiol. 2024 Mar 20;15:1339156. doi: 10.3389/fmicb.2024.1339156. eCollection 2024. Front Microbiol. 2024. PMID: 38572227 Free PMC article. - Improving somatic exome sequencing performance by biological replicates.
Cebeci YE, Erturk RA, Ergun MA, Baysan M. Cebeci YE, et al. BMC Bioinformatics. 2024 Mar 22;25(1):124. doi: 10.1186/s12859-024-05742-5. BMC Bioinformatics. 2024. PMID: 38519906 Free PMC article.
References
- Dalca AV, Brudno M. Genome variation discovery with high-throughput sequencing data. Brief Bioinform. 2010;11:3–14. - PubMed
- Flicek P, Birney E. Sense from sequence reads: methods for alignment and assembly. Nat Methods. 2009;6:S6–12. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous