Patterns of basal signaling heterogeneity can distinguish cellular populations with different drug sensitivities - PubMed (original) (raw)
Patterns of basal signaling heterogeneity can distinguish cellular populations with different drug sensitivities
Dinesh Kumar Singh et al. Mol Syst Biol. 2010.
Abstract
Phenotypic heterogeneity has been widely observed in cellular populations. However, the extent to which heterogeneity contains biologically or clinically important information is not well understood. Here, we investigated whether patterns of basal signaling heterogeneity, in untreated cancer cell populations, could distinguish cellular populations with different drug sensitivities. We modeled cellular heterogeneity as a mixture of stereotyped signaling states, identified based on colocalization patterns of activated signaling molecules from microscopy images. We found that patterns of heterogeneity could be used to separate the most sensitive and resistant populations to paclitaxel within a set of H460 lung cancer clones and within the NCI-60 panel of cancer cell lines, but not for a set of less heterogeneous, immortalized noncancer human bronchial epithelial cell (HBEC) clones. Our results suggest that patterns of signaling heterogeneity, characterized as ensembles of a small number of distinct phenotypic states, can reveal functional differences among cellular populations.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
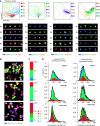
Non-small cell lung cancer H460 clones exhibit a high degree of phenotypic heterogeneity. (A) (Top) Cellular heterogeneity can be characterized as a mixture of phenotypically distinct subpopulations using a Gaussian mixture model (GMM). Shown is the result of computing a ‘reference’ GMM of five subpopulations. Points in GMM scatter plots correspond to individual cells, visualized through feature representation and PCA reduction to two dimensions. Colored ellipses represent covariance 1 s.d. from the mean for each Gaussian cluster (see Supplementary information); cells in this (and all subsequent) scatter plot are colored by the subpopulation of maximum probability. (Bottom) Images of four representative cells from each computed subpopulation are shown. (B) Clones display phenotypically diverse signaling states as measured by activation and colocalization patterns of pSTAT3 and pPTEN immunostaining. Although some clones are phenotypically similar to the parent (e.g. clone 65), others are dramatically dissimilar to the parent (e.g. clone 100). (C) Heterogeneity observed in each clonal population is summarized with a subpopulation profile: a vector estimating the proportion of cells in each subpopulation. (D) Cell populations with similar overall distributions of marker intensities may have dramatically different proportions of subpopulations. Stacked histograms of subpopulation intensities are shown for the parent and two clones. Colors correspond to subpopulations identified in (A). Black outlines correspond to overall histogram; vertical lines indicate population medians. Pseudocolors for images in (A) are specified above the scatter plots. Pseudocolors for images in (B) are: DNA-blue, pSTAT3-green, pPTEN-red. Scale bars: 20 μm. MS1 refers to marker set 1.
Figure 2
Distinct patterns of signaling heterogeneity can be compared across H460 clones. Shown are results computed using marker set 1 (DNA/pSTAT3/pPTEN); pseudocolors for the thumbnail images are as in Figure 1B. At the top are the representative GMM scatter plots from eight clones and the parent (P) culture. Below are thumbnail images of each clone. Yellow/blue heat map shows enrichment/de-enrichment of subpopulations (rows) for all clones (columns). Profiles are computed as the log ratio of clone subpopulation proportions relative to the parent. Clone clustering is determined by hierarchical clustering (dendrogram at bottom). The dendrogram is plotted to produce decreasing average sensitivity to paclitaxel (colored squares above the heat map; Supplementary information). Paclitaxel sensitivity is scored relative to the parent and displayed in red (resistant) and green (sensitive) color scale (gray: paclitaxel-sensitivity scores of clones 33 and 35 are unreliable due to an image-focus problem.) (Supplementary information). Similar results using marker sets 2–4 are shown in Supplementary Figure 6.
Figure 3
Clones with similar patterns of subpopulation profiles have similar drug sensitivities. Clone IDs and relative sensitivities to paclitaxel are as in Figure 2. (A) Paclitaxel-sensitive clones can be separated from nonsensitive clones based on patterns of subpopulation profiles. Multidimensional scaling (MDS) is used to visualize the subpopulation vectors of the H460 populations with respect to the Kullback–Leibler divergence measure (Supplementary information). Solid black squares indicate replicates of parental clone from the seven imaging plates; filled circles indicate clones; gray open circles (clones 33 and 35) indicate unreliable sensitivity scores. (B) The closest three neighbors of a clone tend to have similar drug sensitivities across all markers. Clones are sorted from least to greatest sensitivity to paclitaxel. Heat map indicates the number of nearest neighbors to each clone that are among the top 10-most sensitive (top panel) or resistant (bottom panel) to paclitaxel. (C) Clones of similar paclitaxel sensitivity tend to be phenotypically similar across all four marker sets. Thumbnails of clones (columns) labelled in (A) are shown for all four marker sets (rows). Columns are grouped from left to right by decreasing sensitivity to paclitaxel. Scale bar: 20 μm.
Figure 4
Models of H460 lung cancer heterogeneity can be used to classify sensitivity to paclitaxel for other cancer populations. (A) Accuracies of separating paclitaxel-resistant and -sensitive collections of cell populations based on their subpopulation profiles by a linear SVM (random separation: 50%; perfect separation: 100%). Columns correspond to marker sets; rows correspond to different pairs of sensitive and resistant groups of cell populations. ‘All:’ all populations grouped into either resistant or sensitive classes; ‘Extreme 2N:’ populations only included when in the N-most sensitive or resistant populations. All subpopulations are computed based on H460 reference model. ¶Accuracy not statistically significant (_P_>0.05). †Accuracy not 1 s.d. above the average accuracy over all possible permutations of resistant/sensitive assignments (Supplementary Figure 11). *The least resistant cell line was not used for SVM analysis to create a balanced (four resistant, four sensitive) data set (Supplementary information). (B) Noncancerous HBEC clones display less diversity than the panel of H460 clones. (Top panel) HBEC clones show reduced ranges of drug sensitivities (bottom panel) and dissimilarity among phenotypic profiles compared with H460 clones. Reference model for bottom panel is built by sampling both HBEC and H460 clones; the number of subpopulations is varied from 3 to 14. Error bars are 90% confidence intervals based on bootstrapping (Supplementary information). (C) Drug sensitivity among diverse cancer populations can be separated by subpopulation profiles. The H460 reference model was used to compute subpopulation profiles for nine adherent cell lines with the most extreme GI50 values for paclitaxel within the NCI-60 panel.
Similar articles
- Characterizing heterogeneous cellular responses to perturbations.
Slack MD, Martinez ED, Wu LF, Altschuler SJ. Slack MD, et al. Proc Natl Acad Sci U S A. 2008 Dec 9;105(49):19306-11. doi: 10.1073/pnas.0807038105. Epub 2008 Dec 3. Proc Natl Acad Sci U S A. 2008. PMID: 19052231 Free PMC article. - Mathematical deconvolution uncovers the genetic regulatory signal of cancer cellular heterogeneity on resistance to paclitaxel.
Morilla I, Ranea JA. Morilla I, et al. Mol Genet Genomics. 2017 Aug;292(4):857-869. doi: 10.1007/s00438-017-1316-2. Epub 2017 Apr 6. Mol Genet Genomics. 2017. PMID: 28386641 - Cancer cells undergoing epigenetic transition show short-term resistance and are transformed into cells with medium-term resistance by drug treatment.
Poojan S, Bae SH, Min JW, Lee EY, Song Y, Kim HY, Sim HW, Kang EK, Kim YH, Lee HO, Hong Y, Park WY, Jang H, Hong KM. Poojan S, et al. Exp Mol Med. 2020 Jul;52(7):1102-1115. doi: 10.1038/s12276-020-0464-3. Epub 2020 Jul 13. Exp Mol Med. 2020. PMID: 32661348 Free PMC article. - Roles of tumor heterogeneity in the development of drug resistance: A call for precision therapy.
Wu D, Wang DC, Cheng Y, Qian M, Zhang M, Shen Q, Wang X. Wu D, et al. Semin Cancer Biol. 2017 Feb;42:13-19. doi: 10.1016/j.semcancer.2016.11.006. Epub 2016 Nov 10. Semin Cancer Biol. 2017. PMID: 27840278 Review. - Mathematical models of cell phenotype regulation and reprogramming: Make cancer cells sensitive again!
Wooten DJ, Quaranta V. Wooten DJ, et al. Biochim Biophys Acta Rev Cancer. 2017 Apr;1867(2):167-175. doi: 10.1016/j.bbcan.2017.04.001. Epub 2017 Apr 7. Biochim Biophys Acta Rev Cancer. 2017. PMID: 28396217 Free PMC article. Review.
Cited by
- Nonheritable cellular variability accelerates the evolutionary processes of cancer.
Frank SA, Rosner MR. Frank SA, et al. PLoS Biol. 2012;10(4):e1001296. doi: 10.1371/journal.pbio.1001296. Epub 2012 Apr 3. PLoS Biol. 2012. PMID: 22509130 Free PMC article. - Molecular heterogeneity in malignant peripheral nerve sheath tumors associated with neurofibromatosis type 1.
Thomas L, Mautner VF, Cooper DN, Upadhyaya M. Thomas L, et al. Hum Genomics. 2012 Sep 4;6(1):18. doi: 10.1186/1479-7364-6-18. Hum Genomics. 2012. PMID: 23244685 Free PMC article. - Live-Cell Imaging Shows Uneven Segregation of Extrachromosomal DNA Elements and Transcriptionally Active Extrachromosomal DNA Hubs in Cancer.
Yi E, Gujar AD, Guthrie M, Kim H, Zhao D, Johnson KC, Amin SB, Costa ML, Yu Q, Das S, Jillette N, Clow PA, Cheng AW, Verhaak RGW. Yi E, et al. Cancer Discov. 2022 Feb;12(2):468-483. doi: 10.1158/2159-8290.CD-21-1376. Epub 2021 Nov 24. Cancer Discov. 2022. PMID: 34819316 Free PMC article. - Detecting copy number status and uncovering subclonal markers in heterogeneous tumor biopsies.
Parisi F, Ariyan S, Narayan D, Bacchiocchi A, Hoyt K, Cheng E, Xu F, Li P, Halaban R, Kluger Y. Parisi F, et al. BMC Genomics. 2011 May 11;12:230. doi: 10.1186/1471-2164-12-230. BMC Genomics. 2011. PMID: 21569352 Free PMC article. - ODE constrained mixture modelling: a method for unraveling subpopulation structures and dynamics.
Hasenauer J, Hasenauer C, Hucho T, Theis FJ. Hasenauer J, et al. PLoS Comput Biol. 2014 Jul 3;10(7):e1003686. doi: 10.1371/journal.pcbi.1003686. eCollection 2014 Jul. PLoS Comput Biol. 2014. PMID: 24992156 Free PMC article.
References
- Anderson AR, Weaver AM, Cummings PT, Quaranta V (2006) Tumor morphology and phenotypic evolution driven by selective pressure from the microenvironment. Cell 127: 905–915 - PubMed
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S (2004) Bacterial persistence as a phenotypic switch. Science 305: 1622–1625 - PubMed
- Barre B, Vigneron A, Perkins N, Roninson IB, Gamelin E, Coqueret O (2007) The STAT3 oncogene as a predictive marker of drug resistance. Trends Mol Med 13: 4–11 - PubMed
- Boland MV, Murphy RF (2001) A neural network classifier capable of recognizing the patterns of all major subcellular structures in fluorescence microscope images of HeLa cells. Bioinformatics 17: 1213–1223 - PubMed
- Borg I, Groenen P (1997) Modern Multidimensional Scaling: Theory and Applications. New York: Springer-Verlag
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM081549/GM/NIGMS NIH HHS/United States
- T32 GM007062/GM/NIGMS NIH HHS/United States
- R01 GM085442/GM/NIGMS NIH HHS/United States
- P50 CA070907/CA/NCI NIH HHS/United States
- GM007062/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources