Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses - PubMed (original) (raw)
Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses
Linlin Li et al. J Virol. 2010 Jul.
Abstract
Bats are hosts to a variety of viruses capable of zoonotic transmissions. Because of increased contact between bats, humans, and other animal species, the possibility exists for further cross-species transmissions and ensuing disease outbreaks. We describe here full and partial viral genomes identified using metagenomics in the guano of bats from California and Texas. A total of 34% and 58% of 390,000 sequence reads from bat guano in California and Texas, respectively, were related to eukaryotic viruses, and the largest proportion of those infect insects, reflecting the diet of these insectivorous bats, including members of the viral families Dicistroviridae, Iflaviridae, Tetraviridae, and Nodaviridae and the subfamily Densovirinae. The second largest proportion of virus-related sequences infects plants and fungi, likely reflecting the diet of ingested insects, including members of the viral families Luteoviridae, Secoviridae, Tymoviridae, and Partitiviridae and the genus Sobemovirus. Bat guano viruses related to those infecting mammals comprised the third largest group, including members of the viral families Parvoviridae, Circoviridae, Picornaviridae, Adenoviridae, Poxviridae, Astroviridae, and Coronaviridae. No close relative of known human viral pathogens was identified in these bat populations. Phylogenetic analysis was used to clarify the relationship to known viral taxa of novel sequences detected in bat guano samples, showing that some guano viral sequences fall outside existing taxonomic groups. This initial characterization of the bat guano virome, the first metagenomic analysis of viruses in wild mammals using second-generation sequencing, therefore showed the presence of previously unidentified viral species, genera, and possibly families. Viral metagenomics is a useful tool for genetically characterizing viruses present in animals with the known capability of direct or indirect viral zoonosis to humans.
Figures
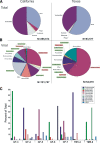
FIG. 1.
Sequence classification for California and Texas bat guano-derived sequences based on BLASTx (E value < 0.001). (A) Percentages of sequences with similarity to those of eukaryotes, bacteria, phages, and eukaryotic virus in GenBank and to unclassifiable sequences. (B) Percentages of most abundant eukaryotic viral matches classified by viral families. Plant viruses are highlighted in green, insect viruses are highlighted in red, and mammalian viruses are not highlighted. (C) Eukaryotic viral families in California roost GF-3 to -7 and in one Texas roost at two collection time points (TM1-4 and TM5-8).
FIG. 2.
Phylogenetic analysis of bat guano-associated nodavirus GF-4 based on its 950-aa RNA-dependent RNA polymerase (RdRp) region. RNA1 of nodaviruses encodes the RdRp protein of ∼1,000 aa.
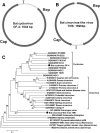
FIG. 3.
(A) Genome organization of bat cyclovirus GF-4; (B) genome organization of bat circovirus-like virus TM6; (C) phylogenetic analysis of bat cyclovirus GF-4 and circovirus-like virus TM6 based on the complete amino acid sequence of the Rep protein (∼280 aa).
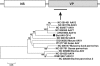
FIG. 4.
Phylogenetic analysis of bat kobuvirus based on its 660-aa partial P1 region. The P1 region of kobuviruses encodes structural proteins of ∼870 aa.
FIG. 5.
Phylogenetic analysis of bat astrovirus based on its 225-aa partial ORF1a region. ORF1a of astroviruses encodes nonstructural protein proteases of ∼900 aa.
FIG. 6.
Phylogenetic analysis of bat parvovirus based on its 210-aa partial nonstructural (NS) ORF. The NS ORF of parvoviruses encodes replication-associated protein REP of ∼700 aa.
FIG. 7.
Phylogenetic analysis of bat AAV based on its 160-aa partial capsid protein VP1 region. The VP1 protein of AAVs is ∼700 aa.
FIG. 8.
Phylogenetic analysis of bat CoV based on its 110-aa partial replicase region. Nonstructural proteins 8 and 9 of CoV are ∼300 aa.
Similar articles
- Virome of Bat Guano from Nine Northern California Roosts.
Li Y, Altan E, Reyes G, Halstead B, Deng X, Delwart E. Li Y, et al. J Virol. 2021 Jan 13;95(3):e01713-20. doi: 10.1128/JVI.01713-20. Print 2021 Jan 13. J Virol. 2021. PMID: 33115864 Free PMC article. - Virome analysis for identification of novel mammalian viruses in bat species from Chinese provinces.
Wu Z, Ren X, Yang L, Hu Y, Yang J, He G, Zhang J, Dong J, Sun L, Du J, Liu L, Xue Y, Wang J, Yang F, Zhang S, Jin Q. Wu Z, et al. J Virol. 2012 Oct;86(20):10999-1012. doi: 10.1128/JVI.01394-12. Epub 2012 Aug 1. J Virol. 2012. PMID: 22855479 Free PMC article. - A metagenomic viral discovery approach identifies potential zoonotic and novel mammalian viruses in Neoromicia bats within South Africa.
Geldenhuys M, Mortlock M, Weyer J, Bezuidt O, Seamark ECJ, Kearney T, Gleasner C, Erkkila TH, Cui H, Markotter W. Geldenhuys M, et al. PLoS One. 2018 Mar 26;13(3):e0194527. doi: 10.1371/journal.pone.0194527. eCollection 2018. PLoS One. 2018. PMID: 29579103 Free PMC article. - [Bats and Viruses: complex relationships].
Rodhain F. Rodhain F. Bull Soc Pathol Exot. 2015 Oct;108(4):272-89. doi: 10.1007/s13149-015-0448-z. Epub 2015 Sep 1. Bull Soc Pathol Exot. 2015. PMID: 26330152 Free PMC article. Review. - Bats as Viral Reservoirs.
Hayman DT. Hayman DT. Annu Rev Virol. 2016 Sep 29;3(1):77-99. doi: 10.1146/annurev-virology-110615-042203. Epub 2016 Aug 22. Annu Rev Virol. 2016. PMID: 27578437 Review.
Cited by
- Myotis fimbriatus Virome, a Window to Virus Diversity and Evolution in the Genus Myotis.
Armero A, Li R, Bienes KM, Chen X, Li J, Xu S, Chen Y, Hughes AC, Berthet N, Wong G. Armero A, et al. Viruses. 2022 Aug 27;14(9):1899. doi: 10.3390/v14091899. Viruses. 2022. PMID: 36146706 Free PMC article. - Virome profiling of bats from Myanmar by metagenomic analysis of tissue samples reveals more novel Mammalian viruses.
He B, Li Z, Yang F, Zheng J, Feng Y, Guo H, Li Y, Wang Y, Su N, Zhang F, Fan Q, Tu C. He B, et al. PLoS One. 2013 Apr 22;8(4):e61950. doi: 10.1371/journal.pone.0061950. Print 2013. PLoS One. 2013. PMID: 23630620 Free PMC article. - High frequency of Aichivirus C (porcine kobuvirus) infection in piglets from different geographic regions of Brazil.
Ribeiro J, de Arruda Leme R, Alfieri AF, Alfieri AA. Ribeiro J, et al. Trop Anim Health Prod. 2013 Nov;45(8):1757-62. doi: 10.1007/s11250-013-0428-x. Epub 2013 Jun 14. Trop Anim Health Prod. 2013. PMID: 23765550 - Detection of kobuvirus RNA in Japanese domestic dogs.
Soma T, Matsubayashi M, Sasai K. Soma T, et al. J Vet Med Sci. 2016 Dec 1;78(11):1731-1735. doi: 10.1292/jvms.16-0217. Epub 2016 Aug 2. J Vet Med Sci. 2016. PMID: 27488907 Free PMC article. - Phylogeny and prevalence of kobuviruses in dogs and cats in the UK.
Carmona-Vicente N, Buesa J, Brown PA, Merga JY, Darby AC, Stavisky J, Sadler L, Gaskell RM, Dawson S, Radford AD. Carmona-Vicente N, et al. Vet Microbiol. 2013 Jun 28;164(3-4):246-52. doi: 10.1016/j.vetmic.2013.02.014. Epub 2013 Feb 26. Vet Microbiol. 2013. PMID: 23490561 Free PMC article.
References
- Blehert, D. S., A. C. Hicks, M. Behr, C. U. Meteyer, B. M. Berlowski-Zier, E. L. Buckles, J. T. Coleman, S. R. Darling, A. Gargas, R. Niver, J. C. Okoniewski, R. J. Rudd, and W. B. Stone. 2009. Bat white-nose syndrome: an emerging fungal pathogen? Science 323:227. - PubMed
- Breitbart, M., M. Haynes, S. Kelley, F. Angly, R. A. Edwards, B. Felts, J. M. Mahaffy, J. Mueller, J. Nulton, S. Rayhawk, B. Rodriguez-Brito, P. Salamon, and F. Rohwer. 2008. Viral diversity and dynamics in an infant gut. Res. Microbiol. 159:367-373. - PubMed
- Breitbart, M., and F. Rohwer. 2005. Method for discovering novel DNA viruses in blood using viral particle selection and shotgun sequencing. Biotechniques 39:729-736. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical