Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies - PubMed (original) (raw)
Review
. 2010 May 14;86(5):749-64.
doi: 10.1016/j.ajhg.2010.04.006.
Margaret P Adam, Swaroop Aradhya, Leslie G Biesecker, Arthur R Brothman, Nigel P Carter, Deanna M Church, John A Crolla, Evan E Eichler, Charles J Epstein, W Andrew Faucett, Lars Feuk, Jan M Friedman, Ada Hamosh, Laird Jackson, Erin B Kaminsky, Klaas Kok, Ian D Krantz, Robert M Kuhn, Charles Lee, James M Ostell, Carla Rosenberg, Stephen W Scherer, Nancy B Spinner, Dimitri J Stavropoulos, James H Tepperberg, Erik C Thorland, Joris R Vermeesch, Darrel J Waggoner, Michael S Watson, Christa Lese Martin, David H Ledbetter
Affiliations
- PMID: 20466091
- PMCID: PMC2869000
- DOI: 10.1016/j.ajhg.2010.04.006
Review
Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies
David T Miller et al. Am J Hum Genet. 2010.
Abstract
Chromosomal microarray (CMA) is increasingly utilized for genetic testing of individuals with unexplained developmental delay/intellectual disability (DD/ID), autism spectrum disorders (ASD), or multiple congenital anomalies (MCA). Performing CMA and G-banded karyotyping on every patient substantially increases the total cost of genetic testing. The International Standard Cytogenomic Array (ISCA) Consortium held two international workshops and conducted a literature review of 33 studies, including 21,698 patients tested by CMA. We provide an evidence-based summary of clinical cytogenetic testing comparing CMA to G-banded karyotyping with respect to technical advantages and limitations, diagnostic yield for various types of chromosomal aberrations, and issues that affect test interpretation. CMA offers a much higher diagnostic yield (15%-20%) for genetic testing of individuals with unexplained DD/ID, ASD, or MCA than a G-banded karyotype ( approximately 3%, excluding Down syndrome and other recognizable chromosomal syndromes), primarily because of its higher sensitivity for submicroscopic deletions and duplications. Truly balanced rearrangements and low-level mosaicism are generally not detectable by arrays, but these are relatively infrequent causes of abnormal phenotypes in this population (<1%). Available evidence strongly supports the use of CMA in place of G-banded karyotyping as the first-tier cytogenetic diagnostic test for patients with DD/ID, ASD, or MCA. G-banded karyotype analysis should be reserved for patients with obvious chromosomal syndromes (e.g., Down syndrome), a family history of chromosomal rearrangement, or a history of multiple miscarriages.
Copyright (c) 2010 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures
Figure 1
Examples of Genomic Imbalances Detected by a CMA but Not by G-Banded Karyotyping (A) A 10.9 Mb deletion, including more than 60 genes. The deletion includes the Williams-Beuren syndrome region at chromosome region 7q11 but extends beyond the typical breakpoints for this syndrome. The arrow is pointing to the deleted chromosome that was observed by retrospective analysis of G-banded slides. (B) A 7.2 Mb duplication on the long arm of chromosome 11. Again, the arrow is pointing to the chromosome that has the duplication shown by the darker G-positive band.
Figure 2
Evolution of a Constitutional CMA Design (A) Early versions of array-based Comparative Genomic Hybridization (aCGH) platforms used for constitutional cytogenetic testing targeted the subtelomeric and pericentromeric regions and defined microdeletion and microduplication syndromes.61,62 (B) Later, more extensive coverage was added at the subtelomeric and pericentromeric regions and included additional probes outside the targeted regions; this is so-called “backbone” coverage. (C) Higher-density backbone coverage or high-density genome-wide arrays provide essentially whole-genome coverage, yielding even higher detection rates.
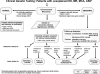
Figure 3
Algorithm for CMA Testing in Patients with Unexplained DD, MR, MCA, and ASD This algorithm assumes that the patient does not present with features of a recognizable syndrome or metabolic disorder or that tests have been negative for a suspected disorder. The first-tier test is a chromosomal copy-number array or CMA. If no copy-number changes are identified, or if only known CNVs that are known to be benign are identified, this testing is considered “normal” (left side of figure), and further clinical evaluation is warranted to determine whether other testing should be pursued on the basis of the clinical presentation. If a CNV is detected within a known, clinically relevant region or gene, or if the CNV is in the genomic backbone and meets recommended size and gene content guidelines, then the result is considered a pathogenic CNV and “abnormal” (right side of figure). For these cases, follow-up analyses include confirmation studies and determination of the mechanism of imbalance in the proband and parental analysis to determine recurrence risk. All other results are considered VOUS until parental analysis is performed to aid in the final clinical interpretation. After the parental analyses of “abnormal” and “VOUS” results, final results may be classified into three major categories: familial variant, abnormal with a low recurrence risk (RR), or abnormal with an increased RR. In addition, the final interpretation may remain VOUS in some instances, even after parental testing.
Similar articles
- Chromosomal Aberrations in Pediatric Patients with Developmental Delay/Intellectual Disability: A Single-Center Clinical Investigation.
Hu T, Zhang Z, Wang J, Li Q, Zhu H, Lai Y, Wang H, Liu S. Hu T, et al. Biomed Res Int. 2019 Nov 6;2019:9352581. doi: 10.1155/2019/9352581. eCollection 2019. Biomed Res Int. 2019. PMID: 31781653 Free PMC article. - Chromosomal Microarray Analysis as a First-Tier Clinical Diagnostic Test in Patients With Developmental Delay/Intellectual Disability, Autism Spectrum Disorders, and Multiple Congenital Anomalies: A Prospective Multicenter Study in Korea.
Jang W, Kim Y, Han E, Park J, Chae H, Kwon A, Choi H, Kim J, Son JO, Lee SJ, Hong BY, Jang DH, Han JY, Lee JH, Kim SY, Lee IG, Sung IK, Moon Y, Kim M, Park JH. Jang W, et al. Ann Lab Med. 2019 May;39(3):299-310. doi: 10.3343/alm.2019.39.3.299. Ann Lab Med. 2019. PMID: 30623622 Free PMC article. - Confirmation of chromosomal microarray as a first-tier clinical diagnostic test for individuals with developmental delay, intellectual disability, autism spectrum disorders and dysmorphic features.
Battaglia A, Doccini V, Bernardini L, Novelli A, Loddo S, Capalbo A, Filippi T, Carey JC. Battaglia A, et al. Eur J Paediatr Neurol. 2013 Nov;17(6):589-99. doi: 10.1016/j.ejpn.2013.04.010. Epub 2013 May 24. Eur J Paediatr Neurol. 2013. PMID: 23711909 - Yield of additional genetic testing after chromosomal microarray for diagnosis of neurodevelopmental disability and congenital anomalies: a clinical practice resource of the American College of Medical Genetics and Genomics (ACMG).
Waggoner D, Wain KE, Dubuc AM, Conlin L, Hickey SE, Lamb AN, Martin CL, Morton CC, Rasmussen K, Schuette JL, Schwartz S, Miller DT; ACMG Professional Practice and Guidelines Committee. Waggoner D, et al. Genet Med. 2018 Oct;20(10):1105-1113. doi: 10.1038/s41436-018-0040-6. Epub 2018 Jun 18. Genet Med. 2018. PMID: 29915380 Free PMC article. Review. - Experience of chromosomal microarray applied in prenatal and postnatal settings in Hong Kong.
Cheng SSW, Chan KYK, Leung KKP, Au PKC, Tam WK, Li SKM, Luk HM, Kan ASY, Chung BHY, Lo IFM, Tang MHY. Cheng SSW, et al. Am J Med Genet C Semin Med Genet. 2019 Jun;181(2):196-207. doi: 10.1002/ajmg.c.31697. Epub 2019 Mar 23. Am J Med Genet C Semin Med Genet. 2019. PMID: 30903683 Review.
Cited by
- Trio-whole exome sequencing reveals the importance of de novo variants in children with intellectual disability and developmental delay.
Li C, Wang Y, Zeng C, Huang B, Chen Y, Xue C, Liu L, Rong S, Lin Y. Li C, et al. Sci Rep. 2024 Nov 11;14(1):27590. doi: 10.1038/s41598-024-79431-x. Sci Rep. 2024. PMID: 39528574 Free PMC article. - Molecular diagnosis of patients with syndromic short stature identified by trio whole-exome sequencing.
Sun H, Zhang G, Li N, Bu X. Sun H, et al. Front Genet. 2024 Oct 2;15:1399186. doi: 10.3389/fgene.2024.1399186. eCollection 2024. Front Genet. 2024. PMID: 39415983 Free PMC article. - Diagnostic and prognostic role of soft ultrasound markers in prenatal detection and assessment of foetal abnormalities.
Moradi B, Bahrami A, Vafaei SM, Sharifpour S, Shariatinia F, Rezvanimehr A, Rashidi-Nezhad A, Fathi M, Yaghoobpoor S, Ghorani H. Moradi B, et al. Prz Menopauzalny. 2024 Jun;23(2):94-108. doi: 10.5114/pm.2024.141092. Epub 2024 Jul 4. Prz Menopauzalny. 2024. PMID: 39391522 Free PMC article. Review. - Survey of the Landscape of Society Practice Guidelines for Genetic Testing of Neurodevelopmental Disorders.
Srivastava S, Cole JJ, Cohen JS, Chopra M, Smith HS, Deardorff MA, Pedapati E, Corner B, Anixt JS, Jeste S, Sahin M, Gurnett CA, Campbell CA; Intellectual and Developmental Disabilities Research Center (IDDRC) Workgroup on Advocating for Access to Genomic Testing. Srivastava S, et al. Ann Neurol. 2024 Nov;96(5):900-913. doi: 10.1002/ana.27045. Epub 2024 Sep 25. Ann Neurol. 2024. PMID: 39319594 Review. - Diagnostic Utility of Whole Genome Sequencing After Negative Karyotyping/Chromosomal Microarray in Infants Born With Multiple Congenital Anomalies.
Yang M, Kim JA, Jo HS, Park JH, Ahn SY, Sung SI, Park WS, Cho HW, Kim JM, Park MH, Park HY, Jang JH, Chang YS. Yang M, et al. J Korean Med Sci. 2024 Sep 23;39(36):e250. doi: 10.3346/jkms.2024.39.e250. J Korean Med Sci. 2024. PMID: 39315442 Free PMC article.
References
- Shevell M., Ashwal S., Donley D., Flint J., Gingold M., Hirtz D., Majnemer A., Noetzel M., Sheth R.D. Practice parameter: evaluation of the child with global developmental delay: Report of the Quality Standards Subcommittee of the American Academy of Neurology and The Practice Committee of the Child Neurology Society. Neurology. 2003;60:367–380. - PubMed
- Autism and Developmental Monitoring Network Surveillance Year 2000 Principal Investigators Prevalence of autism spectrum disorders–autism and developmental disabilities monitoring network, six sites, United States, 2000. MMWR Surveill. Summ. 2007;56:1–11. - PubMed
- Newschaffer C.J., Croen L.A., Daniels J., Giarelli E., Grether J.K., Levy S.E., Mandell D.S., Miller L.A., Pinto-Martin J., Reaven J. The epidemiology of autism spectrum disorders. Annu. Rev. Public Health. 2007;28:235–258. - PubMed
- Moeschler J.B., Shevell M. Clinical genetic evaluation of the child with mental retardation or developmental delays. Pediatrics. 2006;117:2304–2316. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 MH074090/MH/NIMH NIH HHS/United States
- RC2 HD064525/HD/NICHD NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- MH074090/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical