MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis - PubMed (original) (raw)
MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis
S Hani Najafi-Shoushtari et al. Science. 2010.
Abstract
Proper coordination of cholesterol biosynthesis and trafficking is essential to human health. The sterol regulatory element-binding proteins (SREBPs) are key transcription regulators of genes involved in cholesterol biosynthesis and uptake. We show here that microRNAs (miR-33a/b) embedded within introns of the SREBP genes target the adenosine triphosphate-binding cassette transporter A1 (ABCA1), an important regulator of high-density lipoprotein (HDL) synthesis and reverse cholesterol transport, for posttranscriptional repression. Antisense inhibition of miR-33 in mouse and human cell lines causes up-regulation of ABCA1 expression and increased cholesterol efflux, and injection of mice on a western-type diet with locked nucleic acid-antisense oligonucleotides results in elevated plasma HDL. Our findings indicate that miR-33 acts in concert with the SREBP host genes to control cholesterol homeostasis and suggest that miR-33 may represent a therapeutic target for ameliorating cardiometabolic diseases.
Figures
Figure 1
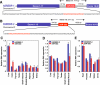
SREBPs are host genes to conserved intronic miRNAs, miR-33a/b, which are co-expressed with SREBPs. Human SREBP-1 (A) and SREBP-2 (B) genes harbor related intronic miRNAs (miR-33b and miR-33a, respectively). The sequences encoding the pri-miRNAs are shown, with the mature miRNA sequences highlighted in red. C-E Expression profile of miR-33a/b and SREBP host genes in selected human tissues. Error bars represent experimental S.D.
Figure 2
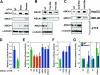
MiR-33 regulates the cholesterol transporter ABCA1. A. Transfection of siRNA against Drosha and Dicer results in increased protein levels of ABCA1 in human HepG2 hepatoma cells (top row), human IMR-90 fibroblasts (middle row), and the mouse macrophage cell line J774 (bottom row). B. Introduction of miR-33a or miR-33b precursors into the three cell lines represses ABCA1 protein levels. C. MiR-33a/b antisense oligonucleotides (A33a/b) increase ABCA1 levels. E. Insertion of the ABCA1 3’UTR fragment into a Luciferase reporter results in decreased Luciferase expression in HEK293 cells. F. Co-transfection with wild-type, but not mutated, miR-33a/b precursors causes further repression of the Luciferase-ABCA1 3’UTR reporter. A scrambled sequence was used as precursor control (PC). G. MiR-33a/b antisense oligonucleotides result in de-repression of the Luciferase-ABCA1 3’UTR reporter. A scrambled sequence was used as antisense control (AC). Error bars represent S.D. * denotes p<0.05.
Figure 3
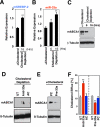
Reciprocal regulation by cholesterol and SREBP/miR-33. Lovastatin/β-cyclodextrin-mediated depletion of cholesterol in J774 mouse macrophages results in increased expression of (A) SREBP-2 and (B) miR-33a, and (C) in decreased ABCA1 protein levels. D. Cholesterol-depleted J774 mouse macrophages exhibit upregulation of ABCA1 protein in response to miR-33a antisense inhibition. E. Transfection of J774 macrophages cultured in the presence of serum/cholesterol with miR-33a precursor results in decreased levels of ABCA1. F. Introduction of miR-33a antisense oligonucleotides into J774 macrophages loaded with radio-labeled cholesterol results in increased cholesterol efflux, whereas miR-33a precursors inhibit cholesterol efflux. Error bars represent S.D. ** denotes p<0.01.
Figure 4
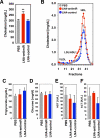
Regulation of HDL by miR-33a in vivo. A. Tail vein injection of mice fed a western-type diet with Locked Nucleic Acid (LNA)-antisense oligonucleotides directed against mouse miR-33a results in elevated total serum cholesterol. B. FPLC analysis of pooled serum from 5 mice on a western-type diet treated revealed an increase in the magnitude of the HDL-cholesterol peak in the LNA-antimiR-treated animals. Elution of lipoprotein standards is indicated by the labels. Plasma triglycerides (C), glucose (D), AST (E), and ALT (F) were unaffected by the treatments. Error bars represent s.e.m. ** denotes p<0.01.
Comment in
- Medicine. HDL miR-ed down by SREBP introns.
Brown MS, Ye J, Goldstein JL. Brown MS, et al. Science. 2010 Jun 18;328(5985):1495-6. doi: 10.1126/science.1192409. Science. 2010. PMID: 20558698 Free PMC article. - Cardiovascular disorders: MicroRNA modulation of cholesterol.
Harrison C. Harrison C. Nat Rev Drug Discov. 2010 Jul;9(7):515. doi: 10.1038/nrd3208. Nat Rev Drug Discov. 2010. PMID: 20592742 No abstract available.
Similar articles
- MiR-33 contributes to the regulation of cholesterol homeostasis.
Rayner KJ, Suárez Y, Dávalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernández-Hernando C. Rayner KJ, et al. Science. 2010 Jun 18;328(5985):1570-3. doi: 10.1126/science.1189862. Epub 2010 May 13. Science. 2010. PMID: 20466885 Free PMC article. - MicroRNAs in metabolism and metabolic diseases.
Rottiers V, Najafi-Shoushtari SH, Kristo F, Gurumurthy S, Zhong L, Li Y, Cohen DE, Gerszten RE, Bardeesy N, Mostoslavsky R, Näär AM. Rottiers V, et al. Cold Spring Harb Symp Quant Biol. 2011;76:225-33. doi: 10.1101/sqb.2011.76.011049. Epub 2011 Dec 12. Cold Spring Harb Symp Quant Biol. 2011. PMID: 22156303 Free PMC article. - Pro-apoptotic miRNA-128-2 modulates ABCA1, ABCG1 and RXRα expression and cholesterol homeostasis.
Adlakha YK, Khanna S, Singh R, Singh VP, Agrawal A, Saini N. Adlakha YK, et al. Cell Death Dis. 2013 Aug 29;4(8):e780. doi: 10.1038/cddis.2013.301. Cell Death Dis. 2013. PMID: 23990020 Free PMC article. - MicroRNAs regulating lipid metabolism in atherogenesis.
Rayner KJ, Fernandez-Hernando C, Moore KJ. Rayner KJ, et al. Thromb Haemost. 2012 Apr;107(4):642-7. doi: 10.1160/TH11-10-0694. Epub 2012 Jan 25. Thromb Haemost. 2012. PMID: 22274626 Free PMC article. Review. - Sphingolipid synthetic pathways are major regulators of lipid homeostasis.
Worgall TS. Worgall TS. Adv Exp Med Biol. 2011;721:139-48. doi: 10.1007/978-1-4614-0650-1_9. Adv Exp Med Biol. 2011. PMID: 21910087 Review.
Cited by
- MicroRNAs and atherosclerosis.
Madrigal-Matute J, Rotllan N, Aranda JF, Fernández-Hernando C. Madrigal-Matute J, et al. Curr Atheroscler Rep. 2013 May;15(5):322. doi: 10.1007/s11883-013-0322-z. Curr Atheroscler Rep. 2013. PMID: 23512606 Free PMC article. Review. - Macrophage Mitochondrial Energy Status Regulates Cholesterol Efflux and Is Enhanced by Anti-miR33 in Atherosclerosis.
Karunakaran D, Thrush AB, Nguyen MA, Richards L, Geoffrion M, Singaravelu R, Ramphos E, Shangari P, Ouimet M, Pezacki JP, Moore KJ, Perisic L, Maegdefessel L, Hedin U, Harper ME, Rayner KJ. Karunakaran D, et al. Circ Res. 2015 Jul 17;117(3):266-78. doi: 10.1161/CIRCRESAHA.117.305624. Epub 2015 May 22. Circ Res. 2015. PMID: 26002865 Free PMC article. - Comparative microRNA expression profiles of cynomolgus monkeys, rat, and human reveal that mir-182 is involved in T2D pathogenic processes.
Zhou J, Meng Y, Tian S, Chen J, Liu M, Zhuo M, Zhang Y, Du H, Wang X. Zhou J, et al. J Diabetes Res. 2014;2014:760397. doi: 10.1155/2014/760397. Epub 2014 Nov 4. J Diabetes Res. 2014. PMID: 25530976 Free PMC article. - Anti-atherosclerosis or No Anti-atherosclerosis: That is the miR-33 question.
Näär AM. Näär AM. Arterioscler Thromb Vasc Biol. 2013 Mar;33(3):447-8. doi: 10.1161/ATVBAHA.112.301021. Arterioscler Thromb Vasc Biol. 2013. PMID: 23407174 Free PMC article. No abstract available. - Comparative microRNA Transcriptomes in Domestic Goats Reveal Acclimatization to High Altitude.
Feng S, Ma J, Long K, Zhang J, Qiu W, Li Y, Jin L, Wang X, Jiang A, Liu L, Xiao W, Li X, Tang Q, Li M. Feng S, et al. Front Genet. 2020 Jul 31;11:809. doi: 10.3389/fgene.2020.00809. eCollection 2020. Front Genet. 2020. PMID: 32849809 Free PMC article.
References
- Maxfield FR, Tabas I. Nature. 2005;438:612. - PubMed
- Moller DE, Kaufman KD. Ann. Rev. Med. 2005;56:45. - PubMed
- Brown MS, Goldstein JL. Cell. 1997;89:331. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM071449/GM/NIGMS NIH HHS/United States
- R21DK084459/DK/NIDDK NIH HHS/United States
- R01 DK056626/DK/NIDDK NIH HHS/United States
- R01DK56626/DK/NIDDK NIH HHS/United States
- R21 DK084459/DK/NIDDK NIH HHS/United States
- R01GM071449/GM/NIGMS NIH HHS/United States
- R01DK48873/DK/NIDDK NIH HHS/United States
- R37 DK048873/DK/NIDDK NIH HHS/United States
- P30 DK034854/DK/NIDDK NIH HHS/United States
- P30 DK34854/DK/NIDDK NIH HHS/United States
- R01 DK048873/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases