HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription - PubMed (original) (raw)
HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription
Nanhai He et al. Mol Cell. 2010.
Abstract
Recruitment of the P-TEFb kinase by HIV-1 Tat to the viral promoter triggers the phosphorylation and escape of RNA polymerase II from promoter-proximal pausing. It is unclear, however, if Tat recruits additional host factors that further stimulate HIV-1 transcription. Using a sequential affinity-purification scheme, we have identified human transcription factors/coactivators AFF4, ENL, AF9, and elongation factor ELL2 as components of the Tat-P-TEFb complex. Through the bridging functions of Tat and AFF4, P-TEFb and ELL2 combine to form a bifunctional elongation complex that greatly activates HIV-1 transcription. Without Tat, AFF4 can mediate the ELL2-P-TEFb interaction, albeit inefficiently. Tat overcomes this limitation by bringing more ELL2 to P-TEFb and stabilizing ELL2 in a process that requires active P-TEFb. The ability of Tat to enable two different classes of elongation factors to cooperate and coordinate their actions on the same polymerase enzyme explains why Tat is such a powerful activator of HIV-1 transcription.
Copyright (c) 2010 Elsevier Inc. All rights reserved.
Figures
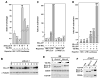
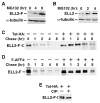
Figure 1. P-TEFb Exists in Two Multisubunit Complexes Containing ELL2/AFF4/Tat/P-TEFb and ELL2/AFF4/P-TEFb, Respectively
(A) CDK9-F, Tat-HA, and their associated factors (lane 1) were isolated through sequential immunoprecipitations (anti-Flag and then anti-HA) from NEs of TTAC-8 cells upon the induction of Tat-HA expression. The immunoprecipitates (IPs) were analyzed on a silver-stained SDS gel, with their identities indicated on the left. NE derived from TTAC-8 cells prior to the induction of Tat-HA was used in a parallel procedure for control (lane 2). MW, molecular weight. (B) The αFlag and αHA IP analyzed in (A) were examined by western blotting for the indicated proteins. (C) The αFlag and αHA sequential IPs derived from NE of HEK293 cells expressing the indicated proteins were analyzedby western blotting as in (B). (D) IPs obtained with the indicated antibodies were examined as in (B). (E) The αFlag IPs derived from NEs of ELL2-F-expressing cells were analyzed as in (B). (F) The parental HEK293 cells and the HEK293-based cell lines expressing the indicated proteins were subjected to anti-Flag immunoprecipitation. The IPs were analyzed by western blotting for the presence of the indicated proteins.
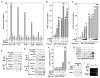
Figure 2. Tat Increases the Levels of ELL2 and the ELL2/P-TEFb-Containing Complex and Synergizes with ELL2 but Not ELL1 to Activate TAR-DependentHIV-1 Transcription
(A) Luciferase activities were measured in extracts of cells cotransfected with the indicated shELL2-expressing constructs, the HIV-1 LTR-luciferase reporter gene, and a vector expressing Tat-HA or nothing. The activity in cells expressing shELL2 #10 but not Tat was set to 1. The error bars represent mean ± SD. (B) Western analyses of the levels of ELL2-F and α-tubulin in cells transfected with the indicated shELL2-expressing constructs. (C) Luciferase activities were measured as in (A) in cells transfected with either WT HIV-1 LTR-luciferase reporter or a ΔTAR mutant construct, together with the indicated plasmids expressing ELL2-F (0.5 ug/well) and/or Tat-HA (0.01 ug/well). (D) Luciferase activities were measured as in (A) in cells transfected with the HIV-1 LTR-luciferase reporter and the indicated ELL1-F-, ELL2-F-, and/or Tat-HA-expressing plasmids. (E) Western analysis of the indicated proteins in NE of cells cotransfected with the indicated cDNA constructs or an empty vector (vec. or “−”). Tat-HA was transfected in 2-fold increments. (F) ELL1-F, ELL2-F, and their associated CDK9 were isolated by anti-Flag IP from NE as analyzed in (E) and examined by western blotting.
Figure 3. Ectopically Expressed AFF4 Acts Like Tat to Synergize with ELL2 to Stimulate Transcriptional Elongation through Promoting ELL2 Accumulation and Association with P-TEFb
(A) Luciferase activities were measured in extracts of cells transfected with the indicated promoter-luciferase constructs together with the ELL2-F- or/and F-AFF4-expressing plasmids as indicated. For each promoter construct, the level of activity detected in the absence of ELL2-F and F-AFF4 was set to 1. The error bars represent mean ± SD. (B and C) Luciferase activities were measured and analyzed as in (A) in cells transfected with the indicated reporter and cDNA constructs. (D and E) NEs of cells transfected with the indicated cDNA constructs (top panel) and anti-HA immunoprecipitates (IP) derived from NE (bottom panel) were analyzed by western blotting (WB) for the indicated proteins. (F) Luciferase reporter assay was performed as in (A) in cells transfected with the indicated reporter and cDNA constructs. (G) Extracts from the same cells as analyzed in (F) were examined by western blotting for the indicated proteins. (H) Transcription reactions containing CDK9-depleted NE, template HIV-1 LTR-G400, and the indicated purified proteins were performed. The 400 nt RNA transcribed from a G-less cassette located at ~1 kb downstream of the HIV-1 promoter is indicated. (I) mRNAs isolated from cells as analyzed in (A) were subjected to RT-PCR analysis with primers that amplify the two indicated regions. RT, reverse transcriptase.
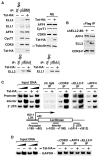
Figure 4. AFF4 Bridges the ELL2-P-TEFb Interaction and Is Required for Stable Accumulation of ELL2
(A) Luciferase activities were measured in cells transfected with the HIV-1 LTR-luciferase reporter gene and constructs expressing shAFF4 or/and Tat-HA. The level of activity detected in the absence of shAFF4 and Tat-HA was set to 1. The error bars represent mean ± SD. (B) NEs (left panel) of HEK293 cells either harboring an empty vector (−) or stably expressing shAFF4 and IP (right panel) obtained from NE with the indicated antibodies were examined by western blotting for the presence of the indicated proteins. (C) NE and anti-Flag IP derived from cells transfected with an empty vector (−) or the construct expressing either the full-length (FL) or truncated (Δ1-300) F-AFF4 were analyzed by western blotting for the indicated proteins. (D) The indicated proteins were incubated with immobilized CycT1-HA/CDK9 in vitro, and the bound proteins were eluted and analyzed by western blotting (left and middle panels). Ten percent of the input FL and Δ1-300 F-AFF4 proteins were examined by anti-Flag western blotting (right panel).
Figure 5. Tat Increases the Amount of ELL2 Bound to P-TEFb, Leading to an Enhanced Association of the ELL2/AFF4/P-TEFb-Containing Complex with the HIV-1 Chromatin Template
(A) NEs from HEK293 cells infected with retroviruses either expressing (+) or not (−) Tat-HA were subjected to western blotting with the indicated antibodies (top right) and immunoprecipitation with anti-CDK9 (top left), anti-ELL2 (bottom left), anti-ELL1 antibodies (bottom right), or a nonspecific rabbit IgG. IPs were analyzed by western blotting for the presence of the indicated proteins. (B) Anti-Flag IP derived from cells stably expressing CDK9-F and harboring either an empty vector (−) or the shELL2 #8-expressing plasmid were analyzed by western blotting for the indicated proteins. (C) Chromatin immunoprecipitation (ChIP) with anti-Flag, anti-CDK9, and anti-AFF4 antibodies was performed in cells containing the integrated HIV-1 LTR-luciferase reporter gene and stably expressing ELL2-F. Three regions corresponding to the promoter, interior, and 3′ UTR of the integrated reporter gene (bottom panel) were PCR amplified from the precipitated and purified DNA. Amplified signals from 5% and 10% of the input chromatin were also shown. (D) ChIP assay was performed as in (C) at the GAPDH locus with the indicated antibodies. The region close to the 3′ end of the gene was PCR amplified from the precipitated DNA.
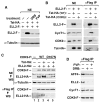
Figure 6. Active P-TEFb Is Required for ELL2 Accumulation and Interaction with P-TEFb
(A) NEs from HEK293 cells transfected with the indicated cDNA constructs and treated with the indicated drugs were analyzed by western blotting for the levels of ELL2-F and α-tubulin. (B and C) NEs (left panel in B and top panel in C) and anti-Flag IP (right panel in B and bottom panel in C) derived from NEs of cells transfected with the indicated cDNA constructs were analyzed by western blotting for the presence of the indicated proteins. (D) F1C2 cells stably expressing CDK9-F were either untreated or treated with the indicated drugs. FVP, flavopiridol. NEs (left panel) and anti-Flag IP (right panel) were analyzed by western blotting.
Figure 7. ELL2 Is a Short-Lived Protein Whose Stability Can Be Significantly Enhanced by Tat or AFF4
(A and B) HEK293 cells containing an ELL2-F-expressing vector (A) or nothing (B) were treated with MG132 for the indicated periods of time. ELL2-F and its endogenous counterpart were detected by anti-Flag (A) and anti-ELL2 (B) western blotting, with α-tubulin serving as a loading control. (C and D) The ELL2-F-producing cells transfected with either an empty vector (−) or a construct expressing Tat-HA (B) or F-AFF4 (C) were pulse labeled with 35S-labeled methionine and L-cysteine and then chased for the indicated periods of time. ELL2-F was then immunoprecipitated and analyzed by SDS-PAGE followed by autoradiography. (E) ELL2-F affinity purified from cells coexpressing Tat-HA were incubated with calf intestine phosphatase (CIP) and analyzed by anti-Flag western blotting.
Similar articles
- HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP.
Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, Benkirane M. Sobhian B, et al. Mol Cell. 2010 May 14;38(3):439-51. doi: 10.1016/j.molcel.2010.04.012. Mol Cell. 2010. PMID: 20471949 Free PMC article. - Super elongation complex promotes early HIV transcription and its function is modulated by P-TEFb.
Kuzmina A, Krasnopolsky S, Taube R. Kuzmina A, et al. Transcription. 2017 May 27;8(3):133-149. doi: 10.1080/21541264.2017.1295831. Epub 2017 Feb 17. Transcription. 2017. PMID: 28340332 Free PMC article. - The AFF4 scaffold binds human P-TEFb adjacent to HIV Tat.
Schulze-Gahmen U, Upton H, Birnberg A, Bao K, Chou S, Krogan NJ, Zhou Q, Alber T. Schulze-Gahmen U, et al. Elife. 2013 Mar 5;2:e00327. doi: 10.7554/eLife.00327. Elife. 2013. PMID: 23471103 Free PMC article. - New insights into the control of HIV-1 transcription: when Tat meets the 7SK snRNP and super elongation complex (SEC).
He N, Zhou Q. He N, et al. J Neuroimmune Pharmacol. 2011 Jun;6(2):260-8. doi: 10.1007/s11481-011-9267-6. Epub 2011 Mar 1. J Neuroimmune Pharmacol. 2011. PMID: 21360054 Free PMC article. Review. - HIV Tat/P-TEFb Interaction: A Potential Target for Novel Anti-HIV Therapies.
Asamitsu K, Fujinaga K, Okamoto T. Asamitsu K, et al. Molecules. 2018 Apr 17;23(4):933. doi: 10.3390/molecules23040933. Molecules. 2018. PMID: 29673219 Free PMC article. Review.
Cited by
- Gene target specificity of the Super Elongation Complex (SEC) family: how HIV-1 Tat employs selected SEC members to activate viral transcription.
Lu H, Li Z, Zhang W, Schulze-Gahmen U, Xue Y, Zhou Q. Lu H, et al. Nucleic Acids Res. 2015 Jul 13;43(12):5868-79. doi: 10.1093/nar/gkv541. Epub 2015 May 24. Nucleic Acids Res. 2015. PMID: 26007649 Free PMC article. - The ubiquitin ligase Siah1 controls ELL2 stability and formation of super elongation complexes to modulate gene transcription.
Liu M, Hsu J, Chan C, Li Z, Zhou Q. Liu M, et al. Mol Cell. 2012 May 11;46(3):325-34. doi: 10.1016/j.molcel.2012.03.007. Epub 2012 Apr 5. Mol Cell. 2012. PMID: 22483617 Free PMC article. - PRMT2 promotes HIV-1 latency by preventing nucleolar exit and phase separation of Tat into the Super Elongation Complex.
Jin J, Bai H, Yan H, Deng T, Li T, Xiao R, Fan L, Bai X, Ning H, Liu Z, Zhang K, Wu X, Liang K, Ma P, Gao X, Hu D. Jin J, et al. Nat Commun. 2023 Nov 10;14(1):7274. doi: 10.1038/s41467-023-43060-1. Nat Commun. 2023. PMID: 37949879 Free PMC article. - PARP1's Involvement in RNA Polymerase II Elongation: Pausing and Releasing Regulation through the Integrator and Super Elongation Complex.
Matveeva EA, Dhahri H, Fondufe-Mittendorf Y. Matveeva EA, et al. Cells. 2022 Oct 12;11(20):3202. doi: 10.3390/cells11203202. Cells. 2022. PMID: 36291070 Free PMC article. - The little elongation complex regulates small nuclear RNA transcription.
Smith ER, Lin C, Garrett AS, Thornton J, Mohaghegh N, Hu D, Jackson J, Saraf A, Swanson SK, Seidel C, Florens L, Washburn MP, Eissenberg JC, Shilatifard A. Smith ER, et al. Mol Cell. 2011 Dec 23;44(6):954-65. doi: 10.1016/j.molcel.2011.12.008. Mol Cell. 2011. PMID: 22195968 Free PMC article.
References
- Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16:92–106. - PubMed
- Chao S-H, Price DH. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem. 2001;276:31793–31799. - PubMed
- De Falco G, Bagella L, Claudio PP, De Luca A, Fu Y, Calabretta B, Sala A, Giordano A. Physical interaction between CDK9 and B-Myb results in suppression of B-Myb gene autoregulation. Oncogene. 2000;19:373–379. - PubMed
- Eissenberg JC, Shilatifard A, Dorokhov N, Michener DE. Cdk9 is an essential kinase in Drosophila that is required for heat shock gene expression, histone methylation and elongation factor recruitment. Mol Genet Genomics. 2007;277:101–114. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI041757/AI/NIAID NIH HHS/United States
- P50 GM082250-03/GM/NIGMS NIH HHS/United States
- R01AI41757-11S1/AI/NIAID NIH HHS/United States
- P50 GM82250/GM/NIGMS NIH HHS/United States
- P50 GM082250/GM/NIGMS NIH HHS/United States
- R01AI41757-11/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous