Animal-to-animal variation in fecal microbial diversity among beef cattle - PubMed (original) (raw)
Animal-to-animal variation in fecal microbial diversity among beef cattle
Lisa M Durso et al. Appl Environ Microbiol. 2010 Jul.
Abstract
The intestinal microbiota of beef cattle are important for animal health, food safety, and methane emissions. This full-length sequencing survey of 11,171 16S rRNA genes reveals animal-to-animal variation in communities that cannot be attributed to breed, gender, diet, age, or weather. Beef communities differ from those of dairy. Core bovine taxa are identified.
Figures
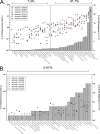
FIG. 1.
Bacterial diversity of six feedlot beef cattle. Gray bars represent the percentages of all 16S sequences that were assigned to each taxonomy. Colored dots represent the percentages of 16S sequences from each library that were assigned to each taxonomic group. Asterisks indicate unclassified members of the named taxon. Panel A shows the data for the first 99% of all the sequences. Panel B shows the data for the remaining 1% of sequences. Note differences in scales for panels A and B.
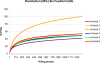
FIG. 2.
Rarefaction curves for six feedlot beef cattle. OTUs were assigned at the 85% DNA sequence similarity level. For comparison purposes, all six curves were truncated after 1,321 sequences.
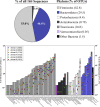
FIG. 3.
Phylum-level distribution of bacterial sequences from six beef feedlot cattle. Asterisks indicate unclassified members of the named taxon.
Similar articles
- A Meta-analysis of Bacterial Diversity in the Feces of Cattle.
Kim M, Wells JE. Kim M, et al. Curr Microbiol. 2016 Feb;72(2):145-151. doi: 10.1007/s00284-015-0931-6. Epub 2015 Nov 6. Curr Microbiol. 2016. PMID: 26542532 - Community structures of fecal bacteria in cattle from different animal feeding operations.
Shanks OC, Kelty CA, Archibeque S, Jenkins M, Newton RJ, McLellan SL, Huse SM, Sogin ML. Shanks OC, et al. Appl Environ Microbiol. 2011 May;77(9):2992-3001. doi: 10.1128/AEM.02988-10. Epub 2011 Mar 4. Appl Environ Microbiol. 2011. PMID: 21378055 Free PMC article. - Virulence-associated and antibiotic resistance genes of microbial populations in cattle feces analyzed using a metagenomic approach.
Durso LM, Harhay GP, Bono JL, Smith TP. Durso LM, et al. J Microbiol Methods. 2011 Feb;84(2):278-82. doi: 10.1016/j.mimet.2010.12.008. Epub 2010 Dec 16. J Microbiol Methods. 2011. PMID: 21167876 - Dysbiosis of the fecal microbiota in feedlot cattle with hemorrhagic diarrhea.
Zeineldin M, Aldridge B, Lowe J. Zeineldin M, et al. Microb Pathog. 2018 Feb;115:123-130. doi: 10.1016/j.micpath.2017.12.059. Epub 2017 Dec 21. Microb Pathog. 2018. PMID: 29275129 - Diversity of the autochthonous colonic microbiota.
Nava GM, Stappenbeck TS. Nava GM, et al. Gut Microbes. 2011 Mar-Apr;2(2):99-104. doi: 10.4161/gmic.2.2.15416. Gut Microbes. 2011. PMID: 21694499 Free PMC article. Review.
Cited by
- Effect of antimicrobial growth promoter administration on the intestinal microbiota of beef cattle.
Reti KL, Thomas MC, Yanke LJ, Selinger LB, Inglis GD. Reti KL, et al. Gut Pathog. 2013 Apr 11;5:8. doi: 10.1186/1757-4749-5-8. eCollection 2013. Gut Pathog. 2013. PMID: 23578222 Free PMC article. - Characterization of Microbial Communities in a Dairy Farm Matrix in Ningxia, China, by 16S rDNA Analysis.
Zhang W, Li W, Ma C, Wu X, Li X, Zeng J, Deng G, Wang Y. Zhang W, et al. Int J Genomics. 2019 Sep 8;2019:3827360. doi: 10.1155/2019/3827360. eCollection 2019. Int J Genomics. 2019. PMID: 31583242 Free PMC article. - Rice straw biochar as a novel niche for improved alterations to the cecal microbial community in rats.
Han J, Meng J, Chen S, Li C, Wang S. Han J, et al. Sci Rep. 2018 Nov 6;8(1):16426. doi: 10.1038/s41598-018-34838-1. Sci Rep. 2018. PMID: 30401962 Free PMC article. - Recto-Anal Junction (RAJ) and Fecal Microbiomes of Cattle Experimentally Challenged With Escherichia coli O157:H7.
Mir RA, Schaut RG, Looft T, Allen HK, Sharma VK, Kudva IT. Mir RA, et al. Front Microbiol. 2020 Apr 17;11:693. doi: 10.3389/fmicb.2020.00693. eCollection 2020. Front Microbiol. 2020. PMID: 32362883 Free PMC article.
References
- Engelbrektson, A., V. Kunin, K. C. Wrighton, N. Zvenigorodsky, F. Chen, H. Ochman, and P. Hugenholtz. 2010. Experimental factors affecting PCR-based estimates of microbial species richness and eveness. ISME J. 4:642-647. - PubMed
- Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164-166.
- Guarner, F., and J. R. Malagelada. 2003. Gut flora in health and disease. Lancet 361:512-519. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases