Mitochondria supply membranes for autophagosome biogenesis during starvation - PubMed (original) (raw)
Mitochondria supply membranes for autophagosome biogenesis during starvation
Dale W Hailey et al. Cell. 2010.
Abstract
Starvation-induced autophagosomes engulf cytosol and/or organelles and deliver them to lysosomes for degradation, thereby resupplying depleted nutrients. Despite advances in understanding the molecular basis of this process, the membrane origin of autophagosomes remains unclear. Here, we demonstrate that, in starved cells, the outer membrane of mitochondria participates in autophagosome biogenesis. The early autophagosomal marker, Atg5, transiently localizes to punctae on mitochondria, followed by the late autophagosomal marker, LC3. The tail-anchor of an outer mitochondrial membrane protein also labels autophagosomes and is sufficient to deliver another outer mitochondrial membrane protein, Fis1, to autophagosomes. The fluorescent lipid NBD-PS (converted to NBD-phosphotidylethanolamine in mitochondria) transfers from mitochondria to autophagosomes. Photobleaching reveals membranes of mitochondria and autophagosomes are transiently shared. Disruption of mitochondria/ER connections by mitofusin2 depletion dramatically impairs starvation-induced autophagy. Mitochondria thus play a central role in starvation-induced autophagy, contributing membrane to autophagosomes.
Copyright (c) 2010 Elsevier Inc. All rights reserved.
Figures
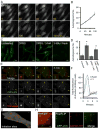
Figure 1. Characterizing the formation of starvation-induced autophagosomes
(A) Time-lapse live-cell imaging of starved NRK58B cells. CFP-LC3 positive structures rapidly proliferated following switch to starvation media. Note concurrent depletion of cytosolic and nuclear pools of CFP-LC3. Growth media (time 0) was replaced with starvation media (subsequent panels). (Scale bar: 20μm) (B) Quantification of CFP-LC3 positive structures. Autophagosomes were counted in sequential time-lapse frames and plotted as a function of time in starvation media. Data points show the mean average of 20 cells; error bars show 1 SD (C) Treatment with Class III PI(3) kinase inhibitor, 3-methyladenine (3-MA). Robust formation of CFP-LC3 structures required activity of the kinase. Identical wells were untreated, starved, or starved in the presence of the 3-MA for 2 h (three left panels). Subsequently, starved cells treated with 3-MA were washed, incubated in DPBS and imaged 2 h later (right-most panel). 3-MA treatment abolished recruitment of CFP-LC3 to membranes and depletion of cytosolic and nuclear pools (2nd from right). Washout of 3-MA restored ability of cells to induce autophagosome formation during starvation. (Scale bar: 20μm) (D) Quantification of 3-MA treatments (mean average of 20 cells +/− 1 SD). Asterisk in bar graphs indicates treatment statistically different from untreated cells by Student t-test: p values < 0.001. (E) Time-lapse live-cell imaging of NRK58B cells expressing YFP-mApg5. During starvation, YFP-mApg5 punctae appeared, and subsequently recruited CFP-LC3 and released YFP-mApg5. Arrows indicate two examples. Inset indicates zoom of autophagosome by lower right arrow. See also supplemental Movie 1. (Scale bars: 2μm in inset; 10μm in panel) (F) Quantification of YFP-mApg5 and CFP-LC3 signals in time-lapse frames. Dramatic accumulation of YFP-mApg5 always preceded CFP-LC3 recruitment. YFP-mApg5 persisted <4 min and abruptly released. (G) Mapping sites of autophagosome formation. The transient appearance of YFP-mApg5 punctae that precedes CFP-LC3 recruitment were scattered throughout the cytosol during starvation. (Scale bar: 5μm) (H) Identifying capture of cytosolic proteins in starvation-induced autophagosomes. Freely diffusing signal was depleted by repetitive photobleaching outside the panel region. This depletion revealed a subpopulation of GAPDH-YFP captured within CFP-LC3 labeled structures. (Scale bar: 2μm).
Figure 2. Characterizing the fate of starvation induced autophagosomes
(A) Visualizing fusion of autophagosomes with lysosomes. NRK58B cells were starved and subsequently labeled with a cell permeant vital lysosomal marker. Live-cell imaging revealed fusion events between autophagosomes and lysosomes that caused accumulation of lysosomal marker and coincident loss of CFP-LC3 signal from the autolysosome (arrow). See also supplemental Movie 2. (B) Visualizing turnover of autophagosomes by photo pulse-labeling and live cell imaging. Following a 2 h starvation, PAGFP-LC3 cells were photoactivated and depleted of cytosolic activated signal to pulse-label an existing population of autophagosomes (Top left panel). Live cell imaging revealed time dependent disappearance of the pulse-labeled population (top panels; see also supplemental Movie 3), which was blocked by addition of chloroquine. (C) Quantification of the lifetime of the structures. Note the turnover of starvation-induced autophagosomes was surprisingly efficient: t1/2 of ~25 minutes.
Figure 3. Evaluating exogenous membrane-targeted markers for transfer to autophagosomes
(A) Expression of chimeric-YFP membrane markers in starvation induced NRK58B cells. Chimeric YFP markers targeting intracellular membrane systems were expressed in NRK58B cells (shown left to right: early endosomal system, Golgi, trans Golgi network, ER, and mitochondrial outer membrane). The mitochondrial outer membrane marker, YFP-Mitocb5TM, uniquely co-localized with induced autophagosomes. (Scale bar: 15μm) (B) Quantification of overlap of CFP-LC3 signal with membrane marker signal. For each marker, autophagosomes with greater than 25% CFP signal overlap with YFP signal were counted; this number was divided by the total number of autophagosomes to determine percent of total autophagosomes that overlapped with marker. The mean average of twenty cells is shown +/− 1 SD. (C) Assessment of the stability of YFP-Mitocb5TM on autophagosomal membranes. YFP-Mitocb5TM-positive autophagosomes were identified. YFP-Mitocb5TM was subsequently bleached in the remainder of the cell (photobleached region indicated by hashed line, top right panel). Unbleached YFP-Mitocb5TM signal persisted on autophagosomes (time series in seconds, lower panel). (Scale bar: 10μm upper panel; 1.5μm lower panel)
Figure 4. Assessing whether mitophagy underlies the appearance of YFP-Mitocb5TM on autophagosomes
(A) Comparison of CFP-LC3 signal overlap with mitochondrial outer membrane (MOM), inner membrane and matrix signal. High-resolution imaging of NRK58B cells expressing MOM, inner membrane, and matrix markers revealed robust overlap of CFP-LC3 signal with the outer membrane, but not the inner membrane and matrix marker (see arrows). (B) Quantification of overlap of CFP-LC3 signal with mitochondrial markers. Quantification was done as described in Fig 3B. (C) Assessment of the association of YFP-Mitocb5TM with the autophagosomal membrane. NRK58B cells were co-transfected with YFP-Mitocb5TM and GAPDH-RFP, and starved. Signal from freely diffusing GAPDH-RFP was depleted by photobleaching to reveal CFP-LC3/YFP-Mitocb5TM/GAPDH-RFP positive autophagosomes. High-resolution images of these structures revealed YFP-Mitocb5TM present on the membrane, not trapped in the lumen. (Scale bar: 1.5μm) (D) Line-scan evaluation of CFP-LC3/YFP-Mitocb5TM/GAPDH-RFP signal in autophagosomes. CFP-LC3 and YFP-Mitocb5TM pixel values along a transecting line (shown in A) exhibited two delineated peaks (membrane). In contrast GAPDH-RFP pixel values along this line exhibited a bell-curve like signal (lumen).
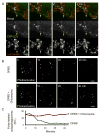
Figure 5. Assessing the association of starvation-induced autophagosomes and mitochondria
(A) Live-cell imaging of GFP-LC3 labeled autophagosomes and associated mitochondrial elements. NRK cells were transfected with the mitochondria matrix marker Mito-RFP and autophagosome marker GFP-LC3. High-resolution high speed imaging of starved cells showed autophagosomes grow during tight association with mitochondrial elements. See also supplementary Movie 4. (B) Electron microscopy of starved cells. Electron micrographs revealed the presence of multilammellar structures tightly associated with mitochondrial elements that exclude mitochondrial matrix. While rare in starved cells, these structures were never observed in unstarved cells. (C) Immuno EM of starved NRK58B cells labeled with gold-conjugated antibodies against CFP revealed clusters of gold particles that were observed tightly associated with proximal mitochondrial elements. (D) Model to demonstrate autophagosomal/mitochondrial membrane association assay. Photobleaching the distal end of a mitochondrial element depletes all YFP-Mitocb5TM signal diffusing throughout the membrane. A YFP-Mitocb5TM positive autophagosome whose membrane is continuous with the MOM also loses YFP-Mitocb5TM signal via diffusion between the two associated organelles. A YFP-Mitocb5TM positive autophagosome that is near but not continuous with the MOM retains YFP-Mitocb5TM signal. (E) Data showing autophagosomal/mitochondrial membrane continuity. Autophagosomes that appeared to be associated with mitochondrial elements were identified. Distal ends of associated mitochondrial elements were targeted with 405nm and 490nm light (yellow box). Here, distal photobleaching depleted signal both from the mitochondria and the associated autophagosome outside the bleached region due to diffusion of the marker from outside the target region into the target region. (see loss of signal, bottom row, middle panel). (Scale bar: 2μm) (F) Autophagosomes that were spatially close but not associated retained signal after photobleaching of proximal mitochondrial elements (see retention of signal, bottom row, middle panel). (Scale bar: 2μm)
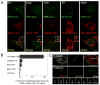
Figure 6. Mitochondrial lipid utilization in autophagosome formation
(A) Time course following loading of exogenous NBD-phosphatidylserine (NBD-PS). NRK cells were exposed to 220 μM NBD-PS, subsequently washed, and maintained in complete media. NBD signal rapidly accumulated in the mitochondria; by 4 hours, the majority of NBD signal was lost. (Scale bar: 5μm) (B) Colocalization of NBD-PS and mitochondria label. High-resolution imaging of Mitotracker red and NBD-PS revealed accumulation of NBD-PS signal in mitochondria one hour after exogenous NBD-PS loading. (C) NBD-labeling of starvation-induced autophagosomes. Cells were were labeled with NBD-PS and allowed 1 hour for NBD to accumulate in mitochondria before starvation. Upon starvation, the NBD-signal accumulated in induced (mCherry-LC3 positive) autophagosomes. (Scale bar: 5μM) (D) Disrupting ER-mitochondria connections by Mitofusin 2 deficiency. Mfn2+/+ and Mfn2−/− MEF cells were transfected with human mCherry-LC3. Mfn2+/+ and Mfn2−/− cells under nutrient rich starvation conditions show a similar degree of basal autophagy. Following starvation, Mfn2−/− cells fail to induce autophagy. Mfn2+/+ cells show a dramatic increase in the number of autophagic structures, as well as depletion of the cytoplasmic and nuclear pools of mcherry-LC3 under starvation conditions. Mfn2−/− cells (like 3-MA treated cells) fail to induce more autophagosomes and clear cytosolic and nuclear mCherry-LC3. (E) Quantification of mCherry-LC3 positive structures in Mfn2+/+ and Mfn2−/− cells. Autophagosomes were counted in 25 cells in three independent experiments.
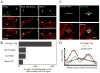
Figure 7. Maintenance of mitochondria during autophagosome biogenesis
(A) YFP-Mitocb5TM uniquely labels autophagosomes; other outer membrane mitochondrial proteins with transmembrane domains that span the entire membrane fail to label autophagosomes. The paucity of other markers supports a unique mechanism for delivery of YFP-Mitocb5TM marker due to its particular membrane association. (B) Schematic of proposed autophagosome bud site. Sharp membrane curvature selects for different lipids on the inner and outer leaflets of curved membranes. The distinct compositions of the leaflets can impede diffusion of proteins with preferences for particular lipid environments. (C) Schematic showing the reported forms of the cb5 transmembrane domain. The wild-type can exist in a kinked helix that interacts only with the outer leaflet of a target membrane. The P115A mutant intercalates across both leaflets of the bilayer. (D) Image showing lack of colocalization of the P115A cb5 mutant and the autophagosomal marker CFP-LC3. (E) Quantitative comparison of the two forms of the outer membrane marker. Mutating Proline115 to Alanine in the cb5™ transmembrane domain abolishes its delivery to autophagosomes. (Scale bar in D: 10 μm). (F) Schematic for one proposed means by which autophagosomes might utilize mitochondrial membrane during autophagosome biogenesis (described in Discussion).
Comment in
- Not all autophagy membranes are created equal.
McEwan DG, Dikic I. McEwan DG, et al. Cell. 2010 May 14;141(4):564-6. doi: 10.1016/j.cell.2010.04.030. Cell. 2010. PMID: 20478247 - Autophagy: From one membrane to another.
Wrighton KH. Wrighton KH. Nat Rev Mol Cell Biol. 2010 Jul;11(7):464. doi: 10.1038/nrm2920. Epub 2010 Jun 9. Nat Rev Mol Cell Biol. 2010. PMID: 20531425 No abstract available.
Similar articles
- Autophagosomes form at ER-mitochondria contact sites.
Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, Amano A, Yoshimori T. Hamasaki M, et al. Nature. 2013 Mar 21;495(7441):389-93. doi: 10.1038/nature11910. Epub 2013 Mar 3. Nature. 2013. PMID: 23455425 - Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation.
Rambold AS, Kostelecky B, Elia N, Lippincott-Schwartz J. Rambold AS, et al. Proc Natl Acad Sci U S A. 2011 Jun 21;108(25):10190-5. doi: 10.1073/pnas.1107402108. Epub 2011 Jun 6. Proc Natl Acad Sci U S A. 2011. PMID: 21646527 Free PMC article. - HRES-1/Rab4 promotes the formation of LC3(+) autophagosomes and the accumulation of mitochondria during autophagy.
Talaber G, Miklossy G, Oaks Z, Liu Y, Tooze SA, Chudakov DM, Banki K, Perl A. Talaber G, et al. PLoS One. 2014 Jan 3;9(1):e84392. doi: 10.1371/journal.pone.0084392. eCollection 2014. PLoS One. 2014. PMID: 24404161 Free PMC article. - Mechanisms of autophagosome biogenesis.
Rubinsztein DC, Shpilka T, Elazar Z. Rubinsztein DC, et al. Curr Biol. 2012 Jan 10;22(1):R29-34. doi: 10.1016/j.cub.2011.11.034. Curr Biol. 2012. PMID: 22240478 Review. - Mitochondria: one of the origins for autophagosomal membranes?
Luo S, Chen Q, Cebollero E, Xing D. Luo S, et al. Mitochondrion. 2009 Jul;9(4):227-31. doi: 10.1016/j.mito.2009.04.004. Epub 2009 May 3. Mitochondrion. 2009. PMID: 19398041 Review.
Cited by
- Phospholipid Scramblase Activity of VDAC Dimers: New Implications for Cell Death, Autophagy and Ageing.
Rockenfeller P. Rockenfeller P. Biomolecules. 2024 Sep 26;14(10):1218. doi: 10.3390/biom14101218. Biomolecules. 2024. PMID: 39456151 Free PMC article. Review. - Targeting the Mitochondria-Proteostasis Axis to Delay Aging.
Zimmermann A, Madreiter-Sokolowski C, Stryeck S, Abdellatif M. Zimmermann A, et al. Front Cell Dev Biol. 2021 Mar 11;9:656201. doi: 10.3389/fcell.2021.656201. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33777963 Free PMC article. - IRGM in autophagy and viral infections.
Petkova DS, Viret C, Faure M. Petkova DS, et al. Front Immunol. 2013 Jan 17;3:426. doi: 10.3389/fimmu.2012.00426. eCollection 2012. Front Immunol. 2013. PMID: 23335927 Free PMC article. - Autophagy in immunity: implications in etiology of autoimmune/autoinflammatory diseases.
Zhou XJ, Zhang H. Zhou XJ, et al. Autophagy. 2012 Sep;8(9):1286-99. doi: 10.4161/auto.21212. Epub 2012 Aug 14. Autophagy. 2012. PMID: 22878595 Free PMC article. Review. - Eaten alive: novel insights into autophagy from multicellular model systems.
Zhang H, Baehrecke EH. Zhang H, et al. Trends Cell Biol. 2015 Jul;25(7):376-87. doi: 10.1016/j.tcb.2015.03.001. Epub 2015 Apr 7. Trends Cell Biol. 2015. PMID: 25862458 Free PMC article. Review.
References
- Achleitner G, Gaigg B, Krasser A, Kainersdorfer E, Kohlwein SD, Perktold A, Zellnig G, Daum G. Association between the endoplasmic reticulum and mitochondria of yeast facilitates interorganelle transport of phospholipids through membrane contact. Eur J Biochem. 1999;264:545–53. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous