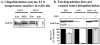Tau protein degradation is catalyzed by the ATP/ubiquitin-independent 20S proteasome under normal cell conditions - PubMed (original) (raw)
Tau protein degradation is catalyzed by the ATP/ubiquitin-independent 20S proteasome under normal cell conditions
Tilman Grune et al. Arch Biochem Biophys. 2010.
Abstract
Tau is the major protein exhibiting intracellular accumulation in Alzheimer disease. The mechanisms leading to its accumulation are not fully understood. It has been proposed that the proteasome is responsible for degrading tau but, since proteasomal inhibitors block both the ubiquitin-dependent 26S proteasome and the ubiqutin-independent 20S proteasome pathways, it is not clear which of these pathways is involved in tau degradation. Some involvement of the ubiquitin ligase, CHIP in tau degradation has also been postulated during stress. In the current studies, we utilized HT22 cells and tau-transfected E36 cells in order to test the relative importance or possible requirement of the ubiquitin-dependent 26S proteasomal system versus the ubiquitin-independent 20S proteasome, in tau degradation. By means of ATP-depletion, ubiquitinylation-deficient E36ts20 cells, a 19S proteasomal regulator subunit MSS1-siRNA approaches, and in vitro ubiquitinylation studies, we were able to demonstrate that ubiquitinylation is not required for normal tau degradation.
2010 Elsevier Inc. All rights reserved.
Figures
Fig. 1. Tau-degradation in HT22 cells is proteasome, but not ATP-dependent
HT22 cells were seeded one day before experiments as described in ‘Materials & Methods’. Cells were incubated in the presence or absence of the proteasome inhibitor, lactacystin (12 μM) for 20h (Panel A). Cells were then lysed and analyzed by immunoblotting using anti-tau and anti-ubiquitin antibodies. One representative out of three experiments is shown. Quantification was performed by using Alpha Ease FC’s “FluorChem 8900”-software. Panel B shows the effect of partial cellular ATP-depletion by KCN and deoxyglucose addition (0.5 mM KCN; 3mM deoxyglucose) on tau levels. The ATP/ADP ratio was calculated from a nucleotide analysis by ion-pair reversed phase HPLC. At the same time point, the content of the 19S proteasomal regulator subunit MSS1, of the tau protein, of GAPDH, and of polyubiquitin conjugates was determined using the corresponding antibodies after immunoblotting. The amounts of human tau and ubiquitin were quantified using AlphaEaseFC’s “FluorChem 8900”-software. Representative results of four independent experiments are shown.
Fig. 2. Tau degradation does not require ubiquitin in transfected ts20 fibroblasts
Human tau40 was transfected into CH E36 and ts20 fibroblasts as described in ‘Materials & Methods.’ The cells were grown at 30.5°C or 39.5°C (restrictive temperature) over night. The content of the E1-enzyme was measured by immunoblot analyses (Panel A). Cycloheximide (‘CHX’) was added at 40 μg × ml−1 and cells were incubated for another 20h. For some experiments cells were incubated in the presence of the proteasome inhibitor, lactacystin (12 μM) for 20h. The tau content was determined by immunoblotting (Panel B). Tau was quantified using AlphaEaseFC’s “FluorChem 8900”-software. Representative results of four independent experiments are shown.
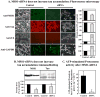
Fig. 3. Tau degradation does not require the complete 26S proteasome: Effects of MSS1-siRNA
HT22 cells were either treated, or not treated, with siRNA directed towards the MSS1 subunit of the 19S regulator: a constitutive component of the 26S proteasome. The exact treatment is extensively described in ‘Materials & Methods.’ Panel A demonstrates the content of MSS1, tau, ubiquitin and GAPDH analyzed by immunofluorescence studies. In parallel a transmitted light and the corresponding immunostained microscopic image are shown. The staining intensity was determined using AlphaEaseFC’s “FluorChem 8900”-software (see right portion of panel A). After the same treatment cells were lysed and analyzed by immunoblotting using anti-MSS1, anti-tau and anti-ubiquitin antibodies (Panel B). Some of the cells were used to determine proteasomal activity. In Panel C the 26S proteasomal activity is shown calculated from the difference of the suc-LLVY-MCA degradation in the presence minus the suc-LLVY-MCA degradation in the absence of ATP. The data presented are Means ± S.E., n=4.
Fig. 4. Tau protein is not ubiquitinylated in vitro
Recombinant human tau40 was prepared and isolated as described in ‘Materials & Methods.’. Ubiquitinylation assays were also performed as described in ‘Materials & Methods,’ using a GST-ubiquitin fusion protein was as described previously [11, 44]. In order to test the electrophoretic and immunoblotting procedure we used recombinant human tau40 as a loading control. Panel A demonstrates the immunoblots of ubiquitinylation assays using lysates of human U87 cells and monitoring either anti-GST-, anti-ubiquitin- or anti-tau-anibodies. In Panel B the same ubiqitinylation assay was performed, with the exception that GAPDH was used as an ubiquitinylation substrate. Consequently an anti-GAPDH-antibody and an anti-ubiquitin antibody were used for detection. Panel C demonstrates immunoblots of the same ubiqitinylation assay with the exception that the ubiqitin activating enzyme E1, the E2 enzyme UbcH5b, the E3-ligase CHIP, and HSC70/HSP40 were used. Both hT40 and htau352 were tested. Analysis was by immonoblotting with anti-GST, anti-tau and anti-TG-5 antibodies. Representative results of four independent experiments are shown.
Fig. 5. Concentration of ubiquitin-conjugates after in vitro ubiquitinylation
The cell lysate mediated ubiquitinylation assay was performed as described in Fig. 4. The resulting protein mix was separated by a S5a-sepharose- (Panel A). The reaction mix of the ubiquitinylation assay was divided into polyubiquitinylated proteins (bound fractions) and non-ubiquitinated proteins (unbound fractions). Analysis was performed by immoblotting using anti-tau and anti-ubiquitin antibodies (Panel A). In further studies the sepharose bound fractions (polyubiquitinated proteins) were either exposed, or not exposed, to ubiquitin hydrolase (0.19μM, for 6h, in 50mM HEPES plus 1mM DTT, at pH 7.5,). The resulting protein mixture was analyzed with anti-tau or anti-ubiquitin antibodies (Panel B) or with anti-TG-5 antibody (Panel C). Isolated hT40 was again used as a loading control to test immunoblot efficiency. An ATP-degrading system, consisting of hexokinase and deoxyglucose; or an ATP-regenerating system, with ATP, phosphoenolpyruvate, pyruvate kinase, and inorganic pyrophosphatase, was used in the ubiquitinylation assay. Representative results of four independent experiments are shown.
Similar articles
- Proteasomal degradation of tau protein.
David DC, Layfield R, Serpell L, Narain Y, Goedert M, Spillantini MG. David DC, et al. J Neurochem. 2002 Oct;83(1):176-85. doi: 10.1046/j.1471-4159.2002.01137.x. J Neurochem. 2002. PMID: 12358741 - Microtubule-associated protein tau is a substrate of ATP/Mg(2+)-dependent proteasome protease system.
Zhang JY, Liu SJ, Li HL, Wang JZ. Zhang JY, et al. J Neural Transm (Vienna). 2005 Apr;112(4):547-55. doi: 10.1007/s00702-004-0196-x. Epub 2004 Sep 14. J Neural Transm (Vienna). 2005. PMID: 15372326 - Degradation of Intrinsically Disordered Proteins by the NADH 26S Proteasome.
Tsvetkov P, Myers N, Adler J, Shaul Y. Tsvetkov P, et al. Biomolecules. 2020 Dec 7;10(12):1642. doi: 10.3390/biom10121642. Biomolecules. 2020. PMID: 33297334 Free PMC article. - Regulating the 20S proteasome ubiquitin-independent degradation pathway.
Ben-Nissan G, Sharon M. Ben-Nissan G, et al. Biomolecules. 2014 Sep 23;4(3):862-84. doi: 10.3390/biom4030862. Biomolecules. 2014. PMID: 25250704 Free PMC article. Review. - [Ubiquitin-independent protein degradation in proteasomes].
Buneeva OA, Medvedev AE. Buneeva OA, et al. Biomed Khim. 2018 Mar;64(2):134-148. doi: 10.18097/PBMC20186402134. Biomed Khim. 2018. PMID: 29723144 Review. Russian.
Cited by
- Small Molecule Modulation of Proteasome Assembly.
Njomen E, Osmulski PA, Jones CL, Gaczynska M, Tepe JJ. Njomen E, et al. Biochemistry. 2018 Jul 17;57(28):4214-4224. doi: 10.1021/acs.biochem.8b00579. Epub 2018 Jun 27. Biochemistry. 2018. PMID: 29897236 Free PMC article. - The proteasome and the degradation of oxidized proteins: Part II - protein oxidation and proteasomal degradation.
Jung T, Höhn A, Grune T. Jung T, et al. Redox Biol. 2014;2:99-104. doi: 10.1016/j.redox.2013.12.008. Epub 2013 Dec 17. Redox Biol. 2014. PMID: 25460724 Free PMC article. Review. - mTOR in Down syndrome: Role in Aß and tau neuropathology and transition to Alzheimer disease-like dementia.
Di Domenico F, Tramutola A, Foppoli C, Head E, Perluigi M, Butterfield DA. Di Domenico F, et al. Free Radic Biol Med. 2018 Jan;114:94-101. doi: 10.1016/j.freeradbiomed.2017.08.009. Epub 2017 Aug 12. Free Radic Biol Med. 2018. PMID: 28807816 Free PMC article. Review. - The Mechanistic Links Between Proteasome Activity, Aging and Age-related Diseases.
Saez I, Vilchez D. Saez I, et al. Curr Genomics. 2014 Feb;15(1):38-51. doi: 10.2174/138920291501140306113344. Curr Genomics. 2014. PMID: 24653662 Free PMC article. - Protein Oxidation in Aging: Does It Play a Role in Aging Progression?
Reeg S, Grune T. Reeg S, et al. Antioxid Redox Signal. 2015 Jul 20;23(3):239-55. doi: 10.1089/ars.2014.6062. Epub 2014 Oct 9. Antioxid Redox Signal. 2015. PMID: 25178482 Free PMC article. Review.
References
- Zhang YJ, Xu YF, Liu YH, Yin J, Li HL, Wang Q, Wang JZ. Peroxynitrite induces Alzheimer-like tau modifications and accumulation in rat brain and its underlying mechanisms. FASEB J. 2006;20:1431–1442. - PubMed
- Goedert M, Spillantini MG, Davies SW. Curr Opin Neurobiol. 1998;8:619–632. - PubMed
- Lee V, Goedert M, Trojanowski JQ. Neurodegenerative tauopathies. Annu Rev Neurosci. 2001;24:1121–1159. - PubMed
- de Vrij FM, Fischer DF, van Leeuwen FW, Hol EM. Protein quality control in Alzheimer’s disease by the ubiquitin proteasome system. Prog Neurobiol. 2004;74:249–270. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 ES003598/ES/NIEHS NIH HHS/United States
- R01 ES003598-22/ES/NIEHS NIH HHS/United States
- R56 ES003598/ES/NIEHS NIH HHS/United States
- ES03598/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources