MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair - PubMed (original) (raw)
. 2010 Jul;30(14):3582-95.
doi: 10.1128/MCB.01476-09. Epub 2010 May 17.
Sairei So, Arun Gupta, Rakesh Kumar, Christelle Cayrou, Nikita Avvakumov, Utpal Bhadra, Raj K Pandita, Matthew H Porteus, David J Chen, Jacques Cote, Tej K Pandita
Affiliations
- PMID: 20479123
- PMCID: PMC2897562
- DOI: 10.1128/MCB.01476-09
MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair
Girdhar G Sharma et al. Mol Cell Biol. 2010 Jul.
Abstract
The human MOF gene encodes a protein that specifically acetylates histone H4 at lysine 16 (H4K16ac). Here we show that reduced levels of H4K16ac correlate with a defective DNA damage response (DDR) and double-strand break (DSB) repair to ionizing radiation (IR). The defect, however, is not due to altered expression of proteins involved in DDR. Abrogation of IR-induced DDR by MOF depletion is inhibited by blocking H4K16ac deacetylation. MOF was found to be associated with the DNA-dependent protein kinase catalytic subunit (DNA-PKcs), a protein involved in nonhomologous end-joining (NHEJ) repair. ATM-dependent IR-induced phosphorylation of DNA-PKcs was also abrogated in MOF-depleted cells. Our data indicate that MOF depletion greatly decreased DNA double-strand break repair by both NHEJ and homologous recombination (HR). In addition, MOF activity was associated with general chromatin upon DNA damage and colocalized with the synaptonemal complex in male meiocytes. We propose that MOF, through H4K16ac (histone code), has a critical role at multiple stages in the cellular DNA damage response and DSB repair.
Figures
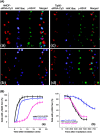
FIG. 1.
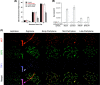
Correlation between status of H4K16ac levels and IR-induced γ-H2AX focus formation in 293 cells. (A) Simultaneous detection of cells transfected with Cy3-labeled siRNA, H4K16ac status, and γ-H2AX focus appearance after exposure to 1.5 Gy of IR. (Aa and Ab) Cells were transfected with hMOF siRNA and analyzed for γ-H2AX foci after 5 min (Aa) and 300 min (Ab) of IR exposure. (Ac and Ad) Cells were transfected with Tip60 siRNA and analyzed for γ-H2AX foci after 5 min (Ac) and 300 min (Ad) of IR exposure. Panels Aa and Ac are magnified cells as shown in Fig. S2A and B in the supplemental material. (B) Frequency of cells with more than 2 γ-H2AX foci observed at various time points postirradiation. For each time point, 100 cells were analyzed. Each experiment was repeated three times; the mean numbers of cells with more than 2 foci are plotted against time. (a) Appearance of γ-H2AX foci in cells with and without depletion of either hMOF or Tip60. (b) Disappearance of γ-H2AX foci in cells with and without depletion of either hMOF or Tip60.
FIG. 2.
Effect of H4K16ac levels on the appearance of IR-induced γ-H2AX foci. (A) 293 cells with and without knockdown of MOF were treated with TSA and examined for H4K16ac levels. Lane C, control. (B) Cells knocked down for hMOF and treated with TSA were irradiated and examined for appearance of γ-H2AX foci. (C) SirT2+/+ and SirT2−/− mouse embryonic fibroblasts with knockdown of MOF, showing MOF and H4K16ac levels and (D) frequency of IR-induced γ-H2AX foci cells. (E) HL60 cells treated with DMSO for different time periods showing MOF, H4K16ac, and histone H4. (F) HL60 cells treated with DMSO or MOF knockdown and irradiated with 1.5 Gy, showing frequency of cells with γ-H2AX postirradiation.
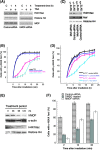
FIG. 3.
hMOF depletion influences IR-induced repair focus-associated proteins and hMOF interaction with chromatin. 293 cells with and without depletion of hMOF with reduced levels of H4K16ac were irradiated with 6 Gy and quantified for foci at different time points postirradiation. (A) For MDC1, cells with more than 5 foci were counted. (B) For 53BP1, cells with more than 3 foci were counted. (C) For induction of hSSB1 foci, cells were irradiated with 2 Gy and cells with more than 2 foci were counted after 1 h postirradiation. (D) RFP-Rad52-expressing 293 cells knocked down for MOF were grown in the presence of bromodeoxyuridine (BrdU) and incubated in Hoechst 33342. Subnuclear DNA damage was induced in the designated rectangular boxes using a 405-nm laser on an LSM510 Zeiss confocal live-imaging system. (E) Association of hMOF with chromatin was determined by ChIP analysis. Cells were irradiated with 5 Gy and fixed, chromatin immunoprecipitation was performed with an hMOF or Rb antibody, and DNA was quantified by absorbance at 260 nm by NanoDrop 2000. (F) Cells with and without exposure to IR were analyzed for the retention of hMOF by Western blotting. hMOF was detected with a specific hMOF antibody.
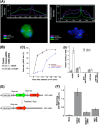
FIG. 4.
MOF depletion results in delayed appearance of γ-H2AX focus at a site-specific DNA DSB. (A) Mouse NIH2/4 cells with and without treatment with TA were fixed and examined for the presence of a γ-H2AX focus. The fluorescence of γ-H2AX, MOF, and DNA was measured across the nucleus and plotted on the top of the cell nucleus. Immunofluorescence was performed with anti-γ-H2AX, anti-H4K16ac, and anti-MOF antibody, and cells were counterstained with DAPI. Fluorescent intensities were measured using Isis or Zen software for each channel. (B) Western blot detection of MOF 72 h posttransfection with siRNA in NIH2/4 cells. (C) NIH2/4 cells with and without MOF depletion were treated with TA and fixed at different time points to detect a γ-H2AX focus by immunostaining. (D) ChIP analysis in I-SceI-induced cells with and without knockdown of MOF examined for bound MOF, H4K16ac, and histone H4. No changes of MOF and H4K16ac levels at the site of damage were detected. (E and F) Effect of MOF on NHEJ. (E) 293 engineered cell line that can become fluorescent (RFP) only upon repair of I-SceI-induced DSBs by NHEJ. CMV, cytomegalovirus; CBA, chicken β-actin promoter. (F) Depletion of MOF results decreased frequency of cells with RFP. These data clearly indicate that the cell's H4K16ac status plays a critical role in the NHEJ pathway.
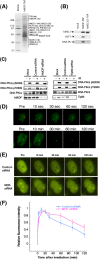
FIG. 5.
Interaction of hMOF with DNA-PKcs. (A) hMOF-TAP is purified with distinct sets of associated proteins. The calmodulin fraction of hMOF-TAP was separated by SDS-PAGE and stained with Sypro Ruby Red, and gel slices were subjected to in-gel digestion and mass spectrometry. M, molecular mass marker. Peptides corresponding to hMSLs and other proteins were identified (numbers of peptides are shown in parentheses) (see Fig. S9 in the supplemental material). DNA-PKcs copurifies preferentially with hMOF. hMOF TAP-, hMSL3L1 TAP-, and mock TAP-purified fractions were assayed for the presence of hMSL1, HCF-1, and DNA-PKcs by Western blotting (B). (C) Cells with and without depletion of hMOF or Tip60 were irradiated with 5 Gy and examined for DNA-PKcs phosphorylation using antibodies specific for pS2056 and pT2609. Depletion of hMOF resulted in loss of ATM-dependent phosphorylation of DNA-PKcs (pT2609), and no such loss was observed in Tip60-depleted cells. (D) Recruitment of DNA-PKcs to the laser-induced DNA DSBs. Cells were transfected with YFP-DNA-PKcs and microirradiated. Time-lapse imaging of YFP-DNA-PKcs-expressing U2OS cells was done before and after microirradiation. (E) Cells stably expressing YFP-DNA-PKcs were treated with control or hMOF siRNA for 72 h, and microirradiated. (F) Initial accumulation kinetics (see Fig. S9B in the supplemental material) and relative fluorescence for a 2-h time course of treatment with YFP-DNA-PKcs at laser-generated DSBs. Error bars represent the standard deviations (SD) from three different experiments.
FIG. 6.
MOF plays a role in HR. (A) Cells with and without MOF or Tip60 knockdown were irradiated with different doses, and cells were quantified for the presence of Rad51 foci after 4 h of irradiation. (B) Impairment of I-SceI-induced HR in hMOF-deficient cells was found. HR frequencies are shown with (+) or without (−) I-SceI induction in untreated cells, in cells treated with control siRNA, and in cells treated with hMOF- or BRCA1-specific siRNA (54). The results presented are the mean and standard error from three independent experiments. (C) Localization of MOF on mouse meiotic chromosomes during prophase 1. Spermatocyte spreads from male mice were immunostained with anti-MOF, anti-TRF1, and anti-SCP. MOF is in red, synaptonemal complex protein SCP3 is green, and TRF1 is white on chromosomes from mouse spermatocytes. The presence of MOF on synaptonemal complexes from leptotene to mid-pachytene suggests the role of MOF in meiotic recombination.
FIG. 7.
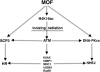
MOF and H4K16ac influence DDR at multiple stages of DNA DSB repair pathways. MOF is a major HAT for H4K16ac, and its levels determine IR-induced repairosome formation. MOF interacts with DNA-PKcs and also localizes on the synaptonemal complex (SCP3) of meiocytes, thus linking to both the DNA DSB repair pathways.
Similar articles
- Bisbenzamidine derivative, pentamidine represses DNA damage response through inhibition of histone H2A acetylation.
Kobayashi J, Kato A, Ota Y, Ohba R, Komatsu K. Kobayashi J, et al. Mol Cancer. 2010 Feb 9;9:34. doi: 10.1186/1476-4598-9-34. Mol Cancer. 2010. PMID: 20144237 Free PMC article. - DNA-PKcs and ATM co-regulate DNA double-strand break repair.
Shrivastav M, Miller CA, De Haro LP, Durant ST, Chen BP, Chen DJ, Nickoloff JA. Shrivastav M, et al. DNA Repair (Amst). 2009 Aug 6;8(8):920-9. doi: 10.1016/j.dnarep.2009.05.006. Epub 2009 Jun 16. DNA Repair (Amst). 2009. PMID: 19535303 Free PMC article. - The impact of heterochromatin on DSB repair.
Goodarzi AA, Noon AT, Jeggo PA. Goodarzi AA, et al. Biochem Soc Trans. 2009 Jun;37(Pt 3):569-76. doi: 10.1042/BST0370569. Biochem Soc Trans. 2009. PMID: 19442252 - The influence of heterochromatin on DNA double strand break repair: Getting the strong, silent type to relax.
Goodarzi AA, Jeggo P, Lobrich M. Goodarzi AA, et al. DNA Repair (Amst). 2010 Dec 10;9(12):1273-82. doi: 10.1016/j.dnarep.2010.09.013. Epub 2010 Oct 30. DNA Repair (Amst). 2010. PMID: 21036673 Review. - Tip60: connecting chromatin to DNA damage signaling.
Sun Y, Jiang X, Price BD. Sun Y, et al. Cell Cycle. 2010 Mar 1;9(5):930-6. doi: 10.4161/cc.9.5.10931. Epub 2010 Mar 11. Cell Cycle. 2010. PMID: 20160506 Free PMC article. Review.
Cited by
- Lamin A/C depletion enhances DNA damage-induced stalled replication fork arrest.
Singh M, Hunt CR, Pandita RK, Kumar R, Yang CR, Horikoshi N, Bachoo R, Serag S, Story MD, Shay JW, Powell SN, Gupta A, Jeffery J, Pandita S, Chen BP, Deckbar D, Löbrich M, Yang Q, Khanna KK, Worman HJ, Pandita TK. Singh M, et al. Mol Cell Biol. 2013 Mar;33(6):1210-22. doi: 10.1128/MCB.01676-12. Epub 2013 Jan 14. Mol Cell Biol. 2013. PMID: 23319047 Free PMC article. - hMOF (human males absent on the first), an oncogenic protein of human oral tongue squamous cell carcinoma, targeting EZH2 (enhancer of zeste homolog 2).
Li Q, Sun H, Shu Y, Zou X, Zhao Y, Ge C. Li Q, et al. Cell Prolif. 2015 Aug;48(4):436-42. doi: 10.1111/cpr.12177. Epub 2015 Jun 1. Cell Prolif. 2015. PMID: 26032517 Free PMC article. - JMJD1C Exhibits Multiple Functions in Epigenetic Regulation during Spermatogenesis.
Nakajima R, Okano H, Noce T. Nakajima R, et al. PLoS One. 2016 Sep 20;11(9):e0163466. doi: 10.1371/journal.pone.0163466. eCollection 2016. PLoS One. 2016. PMID: 27649575 Free PMC article. - Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination.
Gong F, Chiu LY, Cox B, Aymard F, Clouaire T, Leung JW, Cammarata M, Perez M, Agarwal P, Brodbelt JS, Legube G, Miller KM. Gong F, et al. Genes Dev. 2015 Jan 15;29(2):197-211. doi: 10.1101/gad.252189.114. Genes Dev. 2015. PMID: 25593309 Free PMC article. - Epigenetic modifications in double-strand break DNA damage signaling and repair.
Rossetto D, Truman AW, Kron SJ, Côté J. Rossetto D, et al. Clin Cancer Res. 2010 Sep 15;16(18):4543-52. doi: 10.1158/1078-0432.CCR-10-0513. Epub 2010 Sep 7. Clin Cancer Res. 2010. PMID: 20823147 Free PMC article. Review.
References
- Agarwal, M., S. Pandita, C. R. Hunt, A. Gupta, X. Yue, S. Khan, R. K. Pandita, D. Pratt, J. W. Shay, J. S. Taylor, and T. K. Pandita. 2008. Inhibition of telomerase activity enhances hyperthermia-mediated radiosensitization. Cancer Res. 68:3370-3378. - PubMed
- Akhtar, A., and P. B. Becker. 2000. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell 5:367-375. - PubMed
- Allfrey, V. G., B. G. Pogo, V. C. Littau, E. L. Gershey, and A. E. Mirsky. 1968. Histone acetylation in insect chromosomes. Science 159:314-316. - PubMed
- Andegeko, Y., L. Moyal, L. Mittelman, I. Tsarfaty, Y. Shiloh, and G. Rotman. 2001. Nuclear retention of ATM at sites of DNA double strand breaks. J. Biol. Chem. 276:38224-38230. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R13 CA130756/CA/NCI NIH HHS/United States
- R01 CA129537-05/CA/NCI NIH HHS/United States
- R01 CA123232-03/CA/NCI NIH HHS/United States
- R01 CA129537-02/CA/NCI NIH HHS/United States
- R13 CA130756-04/CA/NCI NIH HHS/United States
- R01 NS034746-05S1/NS/NINDS NIH HHS/United States
- R13 CA130756-03/CA/NCI NIH HHS/United States
- R01 CA123232-06/CA/NCI NIH HHS/United States
- R01 CA123232-01A1/CA/NCI NIH HHS/United States
- R13 CA130756-01/CA/NCI NIH HHS/United States
- R01 CA129537/CA/NCI NIH HHS/United States
- R01 CA129537-04/CA/NCI NIH HHS/United States
- R01 NS034746/NS/NINDS NIH HHS/United States
- MOP-64289/CAPMC/ CIHR/Canada
- CA 50519/CA/NCI NIH HHS/United States
- R01 NS034746-06/NS/NINDS NIH HHS/United States
- CA10445/CA/NCI NIH HHS/United States
- R01 CA123232-05/CA/NCI NIH HHS/United States
- R13 CA130756-05/CA/NCI NIH HHS/United States
- R01 CA123232-02/CA/NCI NIH HHS/United States
- CA123232/CA/NCI NIH HHS/United States
- R01 CA123232/CA/NCI NIH HHS/United States
- 64289-2/CAPMC/ CIHR/Canada
- R37 CA050519/CA/NCI NIH HHS/United States
- R01 NS034746-05/NS/NINDS NIH HHS/United States
- R01 NS034746-06S1/NS/NINDS NIH HHS/United States
- R01 CA129537-03/CA/NCI NIH HHS/United States
- R01 CA129537-01A1/CA/NCI NIH HHS/United States
- R01 CA123232-04/CA/NCI NIH HHS/United States
- 87253-1/CAPMC/ CIHR/Canada
- R13 CA130756-02/CA/NCI NIH HHS/United States
- R01 CA050519/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous