Evolution of the sex-related locus and genomic features shared in microsporidia and fungi - PubMed (original) (raw)
Evolution of the sex-related locus and genomic features shared in microsporidia and fungi
Soo Chan Lee et al. PLoS One. 2010.
Abstract
Background: Microsporidia are obligate intracellular, eukaryotic pathogens that infect a wide range of animals from nematodes to humans, and in some cases, protists. The preponderance of evidence as to the origin of the microsporidia reveals a close relationship with the fungi, either within the kingdom or as a sister group to it. Recent phylogenetic studies and gene order analysis suggest that microsporidia share a particularly close evolutionary relationship with the zygomycetes.
Methodology/principal findings: Here we expanded this analysis and also examined a putative sex-locus for variability between microsporidian populations. Whole genome inspection reveals a unique syntenic gene pair (RPS9-RPL21) present in the vast majority of fungi and the microsporidians but not in other eukaryotic lineages. Two other unique gene fusions (glutamyl-prolyl tRNA synthetase and ubiquitin-ribosomal subunit S30) that are present in metazoans, choanoflagellates, and filasterean opisthokonts are unfused in the fungi and microsporidians. One locus previously found to be conserved in many microsporidian genomes is similar to the sex locus of zygomycetes in gene order and architecture. Both sex-related and sex loci harbor TPT, HMG, and RNA helicase genes forming a syntenic gene cluster. We sequenced and analyzed the sex-related locus in 11 different Encephalitozoon cuniculi isolates and the sibling species E. intestinalis (3 isolates) and E. hellem (1 isolate). There was no evidence for an idiomorphic sex-related locus in this Encephalitozoon species sample. According to sequence-based phylogenetic analyses, the TPT and RNA helicase genes flanking the HMG genes are paralogous rather than orthologous between zygomycetes and microsporidians.
Conclusion/significance: The unique genomic hallmarks between microsporidia and fungi are independent of sequence based phylogenetic comparisons and further contribute to define the borders of the fungal kingdom and support the classification of microsporidia as unusual derived fungi. And the sex/sex-related loci appear to have been subject to frequent gene conversion and translocations in microsporidia and zygomycetes.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Unique genome characteristics for the fungi and microsporidia.
The RPS9-RPL21 synteny is uniquely found in the fungal and microsporidian genomes with two exceptions including Schizosaccharomyces species and S. punctatus. The two ribosomal genes, RPS9 and RPL21, are present but unlinked in other non-fungal eukaryotes. Two gene fusions are present in the metazoan and pre-metazoan lineages. Ubiquitin and ribosomal small subunit S30 are encoded by one fused gene (ubiquitin-S30) and two amino-acyl synthetase domains for glutamine and proline are encoded by one gene (glu-pro tRNA synthetase). However, within the opisthokonts, the fungi and microsporidia do not contain either fused gene and two unlinked and separate genes encode each domain.
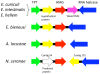
Figure 2. _sex_-related locus of microsporidia.
Encephalitozoon cuniculi, E. intestinalis, and E. hellem share the same _sex_-related locus architecture containing the TPT, HMG, weak HMG, and RNA helicase genes. The weak HMG is not conserved outside Encephalitozoon species examined including E. bieneusi, A. locustae, and N. ceranae. In two insect pathogenic microsporidia, A. locustae and N. ceranae, the RNA helicase gene is unlinked to the HMG genes. The _N. ceranae sex_-related locus contains an additional predicted ORF between the TPT and HMG genes. A hypothetical protein is also linked in the _sex_-related locus across the microsporidia analyzed. Gene sizes are not to scale.
Figure 3. Alignments for HMG and weak HMG proteins in Encephalitozoon species.
Alignment for the HMG proteins shows the HMG gene is highly conserved in eleven E. cuniculi, three E. intestinalis, and one E. hellem isolates suggesting divergent HMG genes are absent in this sample collection in contrast to the zygomycete sex locus which has sexP and sexM alleles. Tandem HMG domains are observed across the three species (highlighted boxes). Alignment for the weak HMG proteins also displays a relatively high level of conservation across the three species.
Figure 4. _sex_-related locus comparison between E. cuniculi isolates.
The _sex_-related locus of EC1 and the reference isolate share 100% sequence identity, whereas the EC2 and EC3 _sex-_related locus has four synonomous (black asterisks) and three nonsynonomous (red asterisks) base substitutions (I38V in the hypothetical protein and C109S and I197M in the RNA helicase). These limited base changes do not support assignment as idiomorphs.
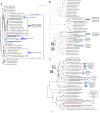
Figure 5. Phylogenetic analyses of the HMG, TPT, and RNA helicases.
(A) One of the E. cuniculi HMG domains (HMG2) is aligned with SexM of the zygomycetes. The other HMG domain (HMG1) is aligned with HMG domains of zygomycetes other than SexP or SexM. The HMG domains of E. bieneusi and A. locustae are also aligned with HMG domains other than SexP or SexM. Note the low bootstrap values on each node of the tree. (B) The microsporidian TPTs are aligned to other TPTs rather than the one in the sex locus. This result suggests that the TPTs in the _sex_-related/sex loci are paralogs (for further discussion, see the text). (C) The microsporidian RNA helicases are also paralogs to the zygomycotan sex locus RNA helicases. Scale indicates an amino acid alteration per position (see the text for further discussion).
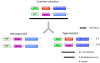
Figure 6. Hypothesis for the paralogous relationship between syntenic genes in zygomycetes and microsporidia.
In the model presented, gene duplication, gene conversion, and chromosome translocation within the syntenic regions resulted in the formation of a syntenic locus with paralogous rather than orthologous genes.
Figure 7. Evolutionary trajectory of the sex locus within zygomycetes and microsporidia.
An ancestral sex locus might have spanned the TPT and RNA helicase genes. Thus, two divergent TPT, HMG, and RNA helicase genes were present in the two _sex_-alleles. In the microsporidian lineage, only one allele was retained. In the zygomycetes, local recombination might have fixed the TPT and RNA helicase genes resulting in one allele for these genes, whereas two alleles of the HMG genes remain in the two opposite mating types.
Similar articles
- Microsporidia evolved from ancestral sexual fungi.
Lee SC, Corradi N, Byrnes EJ 3rd, Torres-Martinez S, Dietrich FS, Keeling PJ, Heitman J. Lee SC, et al. Curr Biol. 2008 Nov 11;18(21):1675-9. doi: 10.1016/j.cub.2008.09.030. Epub 2008 Oct 30. Curr Biol. 2008. PMID: 18976912 Free PMC article. - Zygomycetes, microsporidia, and the evolutionary ancestry of sex determination.
Koestler T, Ebersberger I. Koestler T, et al. Genome Biol Evol. 2011;3:186-94. doi: 10.1093/gbe/evr009. Epub 2011 Feb 9. Genome Biol Evol. 2011. PMID: 21307298 Free PMC article. - Loss of the flagellum happened only once in the fungal lineage: phylogenetic structure of kingdom Fungi inferred from RNA polymerase II subunit genes.
Liu YJ, Hodson MC, Hall BD. Liu YJ, et al. BMC Evol Biol. 2006 Sep 29;6:74. doi: 10.1186/1471-2148-6-74. BMC Evol Biol. 2006. PMID: 17010206 Free PMC article. - Microsporidia: emerging pathogenic protists.
Weiss LM. Weiss LM. Acta Trop. 2001 Feb 23;78(2):89-102. doi: 10.1016/s0001-706x(00)00178-9. Acta Trop. 2001. PMID: 11230819 Review. - Sex in the Mucoralean fungi.
Lee SC, Heitman J. Lee SC, et al. Mycoses. 2014 Dec;57 Suppl 3(0 3):18-24. doi: 10.1111/myc.12244. Epub 2014 Aug 31. Mycoses. 2014. PMID: 25175551 Free PMC article. Review.
Cited by
- Extremely reduced levels of heterozygosity in the vertebrate pathogen Encephalitozoon cuniculi.
Selman M, Sak B, Kváč M, Farinelli L, Weiss LM, Corradi N. Selman M, et al. Eukaryot Cell. 2013 Apr;12(4):496-502. doi: 10.1128/EC.00307-12. Epub 2013 Feb 2. Eukaryot Cell. 2013. PMID: 23376943 Free PMC article. - Dynamics of parasitophorous vacuoles formed by the microsporidian pathogen Encephalitozoon cuniculi.
Lee SC, Heitman J. Lee SC, et al. Fungal Genet Biol. 2017 Oct;107:20-23. doi: 10.1016/j.fgb.2017.07.006. Epub 2017 Jul 25. Fungal Genet Biol. 2017. PMID: 28754285 Free PMC article. - Microsporidia and its relation to Crohn's disease. A retrospective study.
Andreu-Ballester JC, Garcia-Ballesteros C, Amigo V, Ballester F, Gil-Borrás R, Catalán-Serra I, Magnet A, Fenoy S, del Aguila C, Ferrando-Marco J, Cuéllar C. Andreu-Ballester JC, et al. PLoS One. 2013 Apr 18;8(4):e62107. doi: 10.1371/journal.pone.0062107. Print 2013. PLoS One. 2013. PMID: 23637975 Free PMC article. - Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi.
Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W; Fungal Barcoding Consortium; Fungal Barcoding Consortium Author List. Schoch CL, et al. Proc Natl Acad Sci U S A. 2012 Apr 17;109(16):6241-6. doi: 10.1073/pnas.1117018109. Epub 2012 Mar 27. Proc Natl Acad Sci U S A. 2012. PMID: 22454494 Free PMC article. - Population Genetics of Nosema apis and Nosema ceranae: One Host (Apis mellifera) and Two Different Histories.
Maside X, Gómez-Moracho T, Jara L, Martín-Hernández R, De la Rúa P, Higes M, Bartolomé C. Maside X, et al. PLoS One. 2015 Dec 31;10(12):e0145609. doi: 10.1371/journal.pone.0145609. eCollection 2015. PLoS One. 2015. PMID: 26720131 Free PMC article.
References
- Keeling PJ, Fast NM. Microsporidia: biology and evolution of highly reduced intracellular parasites. Annu Rev Microbiol. 2002;56:93–116. - PubMed
- Wittner M, Weiss LM. Washington D.C.: ASM Press; 1999. The microsporidia and microsporidiosis.
- Didier ES, Maddry JA, Brindley PJ, Stovall ME, Didier PJ. Therapeutic strategies for human microsporidia infections. Expert Rev of Anti infec Ther. 2005;3:419–434. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI50113/AI/NIAID NIH HHS/United States
- R21 AI085331/AI/NIAID NIH HHS/United States
- AI052080/AI/NIAID NIH HHS/United States
- T32 AI052080/AI/NIAID NIH HHS/United States
- R01 AI050113/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources