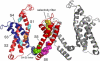Independent and cooperative motions of the Kv1.2 channel: voltage sensing and gating - PubMed (original) (raw)
Independent and cooperative motions of the Kv1.2 channel: voltage sensing and gating
Adva Yeheskel et al. Biophys J. 2010.
Abstract
Voltage-gated potassium (Kv) channels, such as Kv1.2, are involved in the generation and propagation of action potentials. The Kv channel is a homotetramer, and each monomer is composed of a voltage-sensing domain (VSD) and a pore domain (PD). We analyzed the fluctuations of a model structure of Kv1.2 using elastic network models. The analysis suggested a network of coupled fluctuations of eight rigid structural units and seven hinges that may control the transition between the active and inactive states of the channel. For the most part, the network is composed of amino acids that are known to affect channel activity. The results suggested allosteric interactions and cooperativity between the subunits in the coupling between the motion of the VSD and the selectivity filter of the PD, in accordance with recent empirical data. There are no direct contacts between the VSDs of the four subunits, and the contacts between these and the PDs are loose, suggesting that the VSDs are capable of functioning independently. Indeed, they manifest many inherent fluctuations that are decoupled from the rest of the structure. In general, the analysis suggests that the two domains contribute to the channel function both individually and cooperatively.
Copyright 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
Figure 1
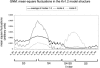
Identification of hinges in the third fluctuation mode. (a) MSF. The minima, corresponding to the hinges, are labeled in green and marked 1–6. (b) Two juxtaposed monomers of Kv1.2's model structure (5): chain A (red) and chain C (gray). Transmembrane helices S1–S6 are marked in red, and the six hinge regions (green) are marked by the gray encircled numbers. The figure was prepared using PyMol (71).
Figure 2
The main hinges as inferred from the eight slowest modes, presented on the 3D model structure of Kv1.2 (5). Only two chains are shown: chain A (left) in colors and chain C (right) in gray. The flexible segment identified in the three slowest modes, containing the S3 and S4 helices and the linker in chain A, is in dark blue. The main chain atoms of the amino acids that were found to serve as hinges are presented as spheres. The hinge in the selectivity filter (T373-D379) is in yellow. The hinge in the S6 helix (I402-V410) is in magenta. The hinge between the linker and the S5 helix (M325-L328) is in green. The hinges in the S1 helix (C181), S2 helix (S217), and S3 helix (V261) are in cyan. These hinges were identified in the first three modes. The hinge in the S4 helix (F302, orange) was identified in the next five modes (4–8). The network of hinges and rigid elements appears to couple between the VSD and PD. The figure was prepared using PyMol (71).
Figure 3
The cooperative motions between residue pairs in the tetramer. The axes mark the residue numbers in each chain. The magnitude of the positive and negative correlations between the fluctuations of the amino acids is color-coded using the red-to-blue scale on the right. (a and d) Interactions within chain A. (b and e) Interactions between residues in chain A and residues in its nearest neighbor (chain B). (c and f) Interactions between residues in chain A and residues in its juxtaposed neighbor (chain C). The different structural segments are marked on the axes. Panels a–c present averages over the three slowest modes, and panels d–f show averages over the next five modes (4–8). A positive correlation indicates a motion of the two residues with the same sense (e.g., in the same direction), whereas a negative correlation indicates a motion with the opposite sense (e.g., in the opposite direction).
Figure 4
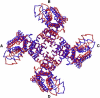
The first ANM mode of fluctuations. The two deformed tetrameric structures, approximately reflecting the motion end-points, are colored blue and red, with the four subunits marked A–D. The original model structure used for the analysis is not presented. The positions of the VSDs (periphery) differ between the two conformations, whereas the PD (center) is, in essence, immobile. The picture was prepared using PyMol (71).
Figure 5
MSF of the Kv1.2 model structure in the five slowest modes. The averaged MSFs of modes 1–3 are in black (identical to Fig. 1_a_), and the fluctuations of the fourth and fifth modes are in dark and light gray, respectively. The hinge in the S4 helix appears only in the fourth and higher modes, in residue F302. Arginine residues (294, 297, 300, and 303) are marked in squares. Only fluctuations of the S3–S5 helices are presented here, even though the calculation was done for the entire structure. A minimum within the S4 helix appears only from the fourth mode.
Figure 6
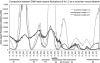
GNM MSF of the tetramer and the monomer. The MSF of the three slowest modes of the monomer are presented in light gray. The MSF of the three slowest modes of the tetramer are in black (identical to Fig. 1_a_) and the next five modes are in dark gray. The locations of the S1–S6 helices are indicated. Most of the hinges identified in the tetramer also appear in the monomer. The main differences are the higher mobility of helix S6 and the shift in the location of the hinge between the domains.
References
- Sanguinetti M.C., Spector P.S. Potassium channelopathies. Neuropharmacology. 1997;36:755–762. - PubMed
- Jiang Y., Lee A., MacKinnon R. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. - PubMed
- Long S.B., Campbell E.B., Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. - PubMed
- Pathak M.M., Yarov-Yarovoy V., Isacoff E.Y. Closing in on the resting state of the Shaker K(+) channel. Neuron. 2007;56:124–140. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous