Rhinovirus genome evolution during experimental human infection - PubMed (original) (raw)
Clinical Trial
Rhinovirus genome evolution during experimental human infection
Samuel Cordey et al. PLoS One. 2010.
Abstract
Human rhinoviruses (HRVs) evolve rapidly due in part to their error-prone RNA polymerase. Knowledge of the diversity of HRV populations emerging during the course of a natural infection is essential and represents a basis for the design of future potential vaccines and antiviral drugs. To evaluate HRV evolution in humans, nasal wash samples were collected daily for five days from 15 immunocompetent volunteers experimentally infected with a reference stock of HRV-39. In parallel, HeLa-OH cells were inoculated to compare HRV evolution in vitro. Nasal wash in vivo assessed by real-time PCR showed a viral load that peaked at 48-72 h. Ultra-deep sequencing was used to compare the low-frequency mutation populations present in the HRV-39 inoculum in two human subjects and one HeLa-OH supernatant collected 5 days post-infection. The analysis revealed hypervariable mutation locations in VP2, VP3, VP1, 2C and 3C genes and conserved regions in VP4, 2A, 2B, 3A, 3B and 3D genes. These results were confirmed by classical sequencing of additional samples, both from inoculated volunteers and independent cell infections, and suggest that HRV inter-host transmission is not associated with a strong bottleneck effect. A specific analysis of the VP1 capsid gene of 15 human cases confirmed the high mutation incidence in this capsid region, but not in the antiviral drug-binding pocket. We could also estimate a mutation frequency in vivo of 3.4x10(-4) mutations/nucleotides and 3.1x10(-4) over the entire ORF and VP1 gene, respectively. In vivo, HRV generate new variants rapidly during the course of an acute infection due to mutations that accumulate in hot spot regions located at the capsid level, as well as in 2C and 3C genes.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
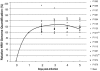
Figure 1. Kinetics of HRV-39 infection in inoculated patients.
The course of infection is shown from days 0 to 5 for each patient (P). The relative HRV RNA values are expressed as 1/CT and converted into % with 100% being arbitrarily set for each patient at day 1. The mean value is represented by the black line. Subjects analyzed further by ultra-deep sequencing are marked with an asterisk.
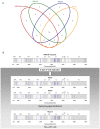
Figure 2. Representation of all specific or common minority mutations to be found in the same loci for the four samples analysed by ultra-deep sequencing.
(A) Venn plot showing the minority mutations present initially in the HRV-39 inoculum and those present in HeLa-A, P1073 and P1077 after 5 days of infection. (B) Minority mutations present in the initial inoculum and in subjects P1073 and P1077 after 5 days of viral replication in the nasopharyngeal epithelium are represented by black and blue at their respective positions along the HRV-39 ORF. Blue bars represent the 14 minority mutations present in the Venn plot in the four viral populations.
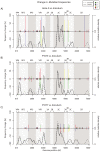
Figure 3. Change in mutation frequencies (minority and majority mutations, colored bars) and minority mutation densities (curves).
Colored bars represent the difference in proportions of each nucleotide between the inoculum and the final (5 days' post-infection) sample in HeLa (A), subjects P1073 (B) and P1077 (C). As each gain in proportion by one nucleotide must be a loss by another, the changes sum to zero. Crosses indicate non-synonymous mutations. Curves indicate minority mutation densities (estimated by a Gaussian kernel function), including mutations whose nucleotide proportions did not change between the inoculum and the final sample.
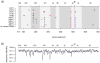
Figure 4. Comparison of ORF consensus sequences in HeLa-OH and human volunteers.
(A) Five HeLa-OH (A to E) and samples from five human volunteers (P1062, P1120, P1073, P1074 and P1077) collected 5 days' post-infection were analysed by classical sequencing method. All mutants present after 5 days of infection, as well as those already present as minority mutations in the initial HRV-39 inoculum, are shown at their respective genome position. Synonymous, non-synonymous and non-conservative mutations are represented by black, and blue and red symbols, respectively. (B) The conservation plot was calculated based on an alignment of 99 rhinovirus serotypes as previously published . The average sequence identity for the ORF was 69.9% (blue dashed line).
Similar articles
- Rhinovirus genome variation during chronic upper and lower respiratory tract infections.
Tapparel C, Cordey S, Junier T, Farinelli L, Van Belle S, Soccal PM, Aubert JD, Zdobnov E, Kaiser L. Tapparel C, et al. PLoS One. 2011;6(6):e21163. doi: 10.1371/journal.pone.0021163. Epub 2011 Jun 21. PLoS One. 2011. PMID: 21713005 Free PMC article. - Analysis of genetic diversity and sites of recombination in human rhinovirus species C.
McIntyre CL, McWilliam Leitch EC, Savolainen-Kopra C, Hovi T, Simmonds P. McIntyre CL, et al. J Virol. 2010 Oct;84(19):10297-310. doi: 10.1128/JVI.00962-10. Epub 2010 Jul 28. J Virol. 2010. PMID: 20668080 Free PMC article. - Genome-wide diversity and selective pressure in the human rhinovirus.
Kistler AL, Webster DR, Rouskin S, Magrini V, Credle JJ, Schnurr DP, Boushey HA, Mardis ER, Li H, DeRisi JL. Kistler AL, et al. Virol J. 2007 May 3;4:40. doi: 10.1186/1743-422X-4-40. Virol J. 2007. PMID: 17477878 Free PMC article. - Proposals for the classification of human rhinovirus species C into genotypically assigned types.
Simmonds P, McIntyre C, Savolainen-Kopra C, Tapparel C, Mackay IM, Hovi T. Simmonds P, et al. J Gen Virol. 2010 Oct;91(Pt 10):2409-19. doi: 10.1099/vir.0.023994-0. Epub 2010 Jul 7. J Gen Virol. 2010. PMID: 20610666 Review. - Analysis of the complete genome sequences of human rhinovirus.
Palmenberg AC, Rathe JA, Liggett SB. Palmenberg AC, et al. J Allergy Clin Immunol. 2010 Jun;125(6):1190-9; quiz 1200-1. doi: 10.1016/j.jaci.2010.04.010. Epub 2010 May 14. J Allergy Clin Immunol. 2010. PMID: 20471068 Free PMC article. Review.
Cited by
- LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets.
Wilm A, Aw PP, Bertrand D, Yeo GH, Ong SH, Wong CH, Khor CC, Petric R, Hibberd ML, Nagarajan N. Wilm A, et al. Nucleic Acids Res. 2012 Dec;40(22):11189-201. doi: 10.1093/nar/gks918. Epub 2012 Oct 12. Nucleic Acids Res. 2012. PMID: 23066108 Free PMC article. - Characterising the mechanism of airway smooth muscle β2 adrenoceptor desensitization by rhinovirus infected bronchial epithelial cells.
Van Ly D, Faiz A, Jenkins C, Crossett B, Black JL, McParland B, Burgess JK, Oliver BG. Van Ly D, et al. PLoS One. 2013;8(2):e56058. doi: 10.1371/journal.pone.0056058. Epub 2013 Feb 15. PLoS One. 2013. PMID: 23457497 Free PMC article. - Within-host rhinovirus evolution in upper and lower respiratory tract highlights capsid variability and mutation-independent compartmentalization.
Makhsous N, Goya S, Avendaño C, Rupp J, Kuypers J, Jerome KR, Boeckh M, Waghmare A, Greninger AL. Makhsous N, et al. bioRxiv [Preprint]. 2023 May 11:2023.05.11.540440. doi: 10.1101/2023.05.11.540440. bioRxiv. 2023. PMID: 37214809 Free PMC article. Updated. Preprint. - Malaria medicines: a glass half full?
Wells TN, Hooft van Huijsduijnen R, Van Voorhis WC. Wells TN, et al. Nat Rev Drug Discov. 2015 Jun;14(6):424-42. doi: 10.1038/nrd4573. Epub 2015 May 22. Nat Rev Drug Discov. 2015. PMID: 26000721 Review. - Effects of rhinovirus species on viral replication and cytokine production.
Nakagome K, Bochkov YA, Ashraf S, Brockman-Schneider RA, Evans MD, Pasic TR, Gern JE. Nakagome K, et al. J Allergy Clin Immunol. 2014 Aug;134(2):332-41. doi: 10.1016/j.jaci.2014.01.029. Epub 2014 Mar 14. J Allergy Clin Immunol. 2014. PMID: 24636084 Free PMC article.
References
- Denny FW., Jr The clinical impact of human respiratory virus infections. Am J Respir Crit Care Med. 1995;152:S4–12. - PubMed
- Kaiser L, Aubert JD, Pache JC, Deffernez C, Rochat T, et al. Chronic rhinoviral infection in lung transplant recipients. Am J Respir Crit Care Med. 2006;174:1392–1399. - PubMed
- Papadopoulos NG, Bates PJ, Bardin PG, Papi A, Leir SH, et al. Rhinoviruses infect the lower airways. J Infect Dis. 2000;181:1875–1884. - PubMed
- Drake JW. The distribution of rates of spontaneous mutation over viruses, prokaryotes, and eukaryotes. Ann N Y Acad Sci. 1999;870:100–107. - PubMed
- Harvala H, Simmonds P. Human parechoviruses: biology, epidemiology and clinical significance. J Clin Virol. 2009;45:1–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources