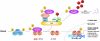Regulation of the Nrf2-Keap1 antioxidant response by the ubiquitin proteasome system: an insight into cullin-ring ubiquitin ligases - PubMed (original) (raw)
Review
. 2010 Dec 1;13(11):1699-712.
doi: 10.1089/ars.2010.3211. Epub 2010 Aug 14.
Affiliations
- PMID: 20486766
- PMCID: PMC2966484
- DOI: 10.1089/ars.2010.3211
Review
Regulation of the Nrf2-Keap1 antioxidant response by the ubiquitin proteasome system: an insight into cullin-ring ubiquitin ligases
Nicole F Villeneuve et al. Antioxid Redox Signal. 2010.
Abstract
Nrf2 is a transcription factor that has emerged as the cell's main defense mechanism against many harmful environmental toxicants and carcinogens. Nrf2 is negatively regulated by Keap1, a substrate adaptor protein for the Cullin3 (Cul3)-containing E3-ligase complex, which targets Nrf2 for ubiquitination and degradation by the ubiquitin proteasome system (UPS). Recent evidence suggests that constitutive activation of Nrf2, due to mutations in Keap1 or Nrf2, is prominent in many cancer types and contributes to chemoresistance. Regulation of Nrf2 by the Cul3-Keap1-E3 ligase provides strong evidence that tight regulation of Cullin-ring ligases (CRLs) is imperative to maintain cellular homeostasis. There are seven known Cullin proteins that form various CRL complexes. They are regulated by neddylation/deneddylation, ubiquitination/deubiquitination, CAND1-assisted complex assembly/disassembly, and subunit dimerization. In this review, we will discuss the regulation of each CRL using the Cul3-Keap1-E3 ligase complex as the primary focus. The substrates of CRLs are involved in many signaling pathways. Therefore, deregulation of CRLs affects several cellular processes, including cell cycle arrest, DNA repair, cell proliferation, senescence, and death, which may lead to many human diseases, including cancer. This makes CRLs a promising target for novel cancer drug therapies.
Figures
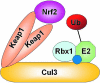
FIG. 1.
A schematic of the Cul3-Keap1-E3 ligase. Cul3 serves as a scaffolding protein that binds the Ub-loaded E2, Rbx1, and Keap1 proteins. Following Nedd8 conjugation (blue circle), a Keap1 homodimer functions as a substrate adaptor to recruit Nrf2 to the Cul3–Keap1–E3 ligase complex for ubiquitination.
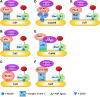
FIG. 2.
A general schematic of the seven Cullin-ring ligases. (A) Cul1, (B) Cul2, (C) Cul3, (D) Cul4A/B = Cul4A and Cul4B, (E) Cul4B, (F) Cul5, and (G) Cul7 Cullin-ring ligases. Words listed in red are all domain names. BPA, BPB, and BPC are the three propeller domains of DDB1.
FIG. 3.
Activation of the NF-κB signaling pathway. Various stimuli can lead to phosphorylation and activation the IKK complex composed of three subunits, IKKα, IKKβ, and IKKγ. Upon activation, IKKβ phosphorylates two serine residues in IκBα that targets it for degradation by the Cul1-β-TrCP1–E3 ligase complex. As a result, the NF-κB complex (p50 and p65) is released, exposing a nuclear localization signal (NLS) that allows the NF-κB complex to translocate to the nucleus and bind to κB sites in the promoters of downstream genes to induce their transcription.
FIG. 4.
Regulation of β-catenin by the Cul1–Skp1–E3 ligase complex. In the absence of Wnt signaling, β-catenin forms a complex with GSK3β, casein kinase 1α (CK1α), Axin, and adenomatous polyposis coli (APC), resulting in CK1α-mediated phosphorylation of β-catenin (S45), followed by GSK3β-mediated phosphorylation of β-catenin at additional residues (T41, S37, S33). Phosphorylation of β-catenin allows it to be recognized by βTrCP1 and targeted for ubiquitination by the Cul1–Skp1–E3 ligase and subsequent degradation by the 26S proteasome.
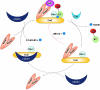
FIG. 5.
Regulation of the Cul3–Keap1–E3 ligase complex. The Cul3–Keap1–E3 ligase complex is active when it is in the neddylated state, which facilitates the docking of a Keap1 homodimer-bound to Nrf2 into the complex, resulting in ubiquitination and degradation of Nrf2. The complex is inactivated by deneddylation. The CSN complex removes Nedd8 from Cul3 enhancing the association of Cul3 and CAND1, which triggers dissociation of Keap1-Nrf2 from Cul3. Finally, neddylation can reactivate the E3 ligase complex. The complex can also be reactivated following deneddylation without disassembly of the complex. UBC12 can conjugate Nedd8 back onto Cul3 without dissociation of Keap1 from the Cul3–Rbx1 core complex (red arrow).
Similar articles
- Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2.
Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Kobayashi A, et al. Mol Cell Biol. 2004 Aug;24(16):7130-9. doi: 10.1128/MCB.24.16.7130-7139.2004. Mol Cell Biol. 2004. PMID: 15282312 Free PMC article. - BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase.
Furukawa M, Xiong Y. Furukawa M, et al. Mol Cell Biol. 2005 Jan;25(1):162-71. doi: 10.1128/MCB.25.1.162-171.2005. Mol Cell Biol. 2005. PMID: 15601839 Free PMC article. - CAND1-mediated substrate adaptor recycling is required for efficient repression of Nrf2 by Keap1.
Lo SC, Hannink M. Lo SC, et al. Mol Cell Biol. 2006 Feb;26(4):1235-44. doi: 10.1128/MCB.26.4.1235-1244.2006. Mol Cell Biol. 2006. PMID: 16449638 Free PMC article. - CRL3s: The BTB-CUL3-RING E3 Ubiquitin Ligases.
Wang P, Song J, Ye D. Wang P, et al. Adv Exp Med Biol. 2020;1217:211-223. doi: 10.1007/978-981-15-1025-0_13. Adv Exp Med Biol. 2020. PMID: 31898230 Review. - The Keap1-Nrf2 system and diabetes mellitus.
Uruno A, Yagishita Y, Yamamoto M. Uruno A, et al. Arch Biochem Biophys. 2015 Jan 15;566:76-84. doi: 10.1016/j.abb.2014.12.012. Epub 2014 Dec 17. Arch Biochem Biophys. 2015. PMID: 25528168 Review.
Cited by
- Activation of Keap1/Nrf2 signaling pathway by nuclear epidermal growth factor receptor in cancer cells.
Huo L, Li CW, Huang TH, Lam YC, Xia W, Tu C, Chang WC, Hsu JL, Lee DF, Nie L, Yamaguchi H, Wang Y, Lang J, Li LY, Chen CH, Mishra L, Hung MC. Huo L, et al. Am J Transl Res. 2014 Nov 22;6(6):649-63. eCollection 2014. Am J Transl Res. 2014. PMID: 25628777 Free PMC article. - Synthesis and Anti-Inflammatory Activity of 2-Amino-4,5,6,7-tetrahydrobenzo[b]thiophene-Derived NRF2 Activators.
Mak KK, Shiming Z, Epemolu O, Dinkova-Kostova AT, Wells G, Gazaryan IG, Sakirolla R, Mohd Z, Pichika MR. Mak KK, et al. ChemistryOpen. 2022 Oct;11(10):e202200181. doi: 10.1002/open.202200181. ChemistryOpen. 2022. PMID: 36284193 Free PMC article. - Differential regulation of proteasome functionality in reproductive vs. somatic tissues of Drosophila during aging or oxidative stress.
Tsakiri EN, Sykiotis GP, Papassideri IS, Gorgoulis VG, Bohmann D, Trougakos IP. Tsakiri EN, et al. FASEB J. 2013 Jun;27(6):2407-20. doi: 10.1096/fj.12-221408. Epub 2013 Mar 1. FASEB J. 2013. PMID: 23457214 Free PMC article. - NRF2 regulates the sensitivity of human NSCLC cells to cystine deprivation-induced ferroptosis via FOCAD-FAK signaling pathway.
Liu P, Wu D, Duan J, Xiao H, Zhou Y, Zhao L, Feng Y. Liu P, et al. Redox Biol. 2020 Oct;37:101702. doi: 10.1016/j.redox.2020.101702. Epub 2020 Aug 27. Redox Biol. 2020. PMID: 32898818 Free PMC article. - Genetic evidence of an evolutionarily conserved role for Nrf2 in the protection against oxidative stress.
Mukaigasa K, Nguyen LT, Li L, Nakajima H, Yamamoto M, Kobayashi M. Mukaigasa K, et al. Mol Cell Biol. 2012 Nov;32(21):4455-61. doi: 10.1128/MCB.00481-12. Epub 2012 Sep 4. Mol Cell Biol. 2012. PMID: 22949501 Free PMC article.
References
- Angers S. Li T. Yi X. MacCoss MJ. Moon RT. Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. - PubMed
- Cardoso F. Ross JS. Picart MJ. Sotiriou C. Durbecq V. Targeting the ubiquitin-proteasome pathway in breast cancer. Clin Breast Cancer. 2004;5:148–157. - PubMed
- Chariot A. The NF-kappaB-independent functions of IKK subunits in immunity and cancer. Trends Cell Biol. 2009;19:404–413. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources