Mouse and zebrafish Hoxa3 orthologues have nonequivalent in vivo protein function - PubMed (original) (raw)
Mouse and zebrafish Hoxa3 orthologues have nonequivalent in vivo protein function
Lizhen Chen et al. Proc Natl Acad Sci U S A. 2010.
Abstract
Hox genes play evolutionarily conserved roles in specifying axial position during embryogenesis. A prevailing paradigm is that changes in Hox gene expression drive evolution of metazoan body plans. Conservation of Hox function across species, and among paralogous Hox genes within a species, supports a model of functional equivalence. In this report, we demonstrate that zebrafish hoxa3a (zfhoxa3a) expressed from the mouse Hoxa3 locus can substitute for mouse Hoxa3 in some tissues, but has distinct or null phenotypes in others. We further show, by using an allele encoding a chimeric protein, that this difference maps primarily to the zfhoxa3a C-terminal domain. Our data imply that the mouse and zebrafish proteins have diverged considerably since their last common ancestor, and that the major difference between them resides in the C-terminal domain. Our data further show that Hox protein function can evolve independently in different cell types or for specific functions. The inability of zfhoxa3a to perform all of the normal roles of mouse Hoxa3 illustrates that Hox orthologues are not always functionally interchangeable.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Structure and expression of Hoxa3 alleles. (A) Scheme of Hoxa3 WT (+), Hoxa3zf (zf), and Hoxa3mz (mz) alleles. Horizontal thin lines represent noncoding genomic DNA at the mouse Hoxa3 locus. Boxes represent exons as follows: gray, 5′ or 3′UTR of mouse Hoxa3; white, mouse Hoxa3 coding sequences; black, zebrafish hoxa3a coding sequences. N, NotI; B, BamHI; S, SpeI. (B) Quantitative RT-PCR shows equivalent mRNA levels for the zfhoxa3a (zf) and mouse Hoxa3 (WT) transcripts in individual whole heterozygous embryos (+/zf 1–3), or in homozygotes (zf/zf, +/+). Whole mount (C and D) and coronal paraffin section (E and F) in situ hybridization of embryonic d 10.5 embryos using allele-specific probes shows identical spatial expression patterns for the WT murine and zf alleles. (G_–_I) Immunofluorescence detection of the HA tag in the zfHoxa3 protein in the hindbrain at embryonic d 10.5. Box in H corresponds to panel in I. Arrow in I shows anterior limit of protein detection. (Scale bars: 1 mm in C and D; 200 μm in G_–_I.)
Fig. 2.
Zebrafish hoxa3a can substitute for mouse Hoxa3 in thyroid, ultimobranchial body, tracheal epithelium, and soft palate development. Transverse (A_–_L; dorsal is up) or sagittal (M_–_P; anterior is up, dorsal is to the left.) paraffin sections of newborn animals; genotypes apply to each column. Scale bars apply to each row. (A_–_D) The thyroid isthmus (arrow) is deleted in Hoxa3null/null (−/−) mice (B), but restored in Hoxa3zf/zf (zf/zf) (C) and Hoxa3mz/mz (mz/mz) (D). (E_–_H) Transverse sections of newborn mice stained with anticalcitonin antibody (brown). Integration of ultimobranchial body–derived C cells is restored in zf/zf (G) and mz/mz (H) mice. (I_–_L) The disorganized tracheal epithelium in the _Hoxa3_-null mutant (J) was not seen in zf/zf (K) or mz/mz (L) animals. The white bar in each panel shows the thickness of the WT epithelium, contrasted with the null mutant (long bar in J). (M_–_P) The posterior palate (velum) is shortened in _Hoxa3_-null mutants (N), but is normal in zf/zf and mz/mz mice (M, O, and P). tr, trachea; es, esophagus; pa, palate; to, tongue. (Scale bars: 200 μm in A_–_D; 100 μm in E_–_H; 50 μm in I_–_L; 800 μm in M_–_P.)
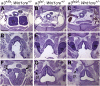
Fig. 3.
Cranial nerve, thymus, and parathyroid defects are not rescued by zfhoxa3a. (A_–_D) Whole-mount antineurofilament staining of embryonic d 10.5 control, Hoxa3null/null (−/−), Hoxa3zf/zf (zf/zf), and Hoxa3mz/mz (mz/mz) embryos. In the control, the IX cranial nerve is connected to hindbrain. (B) In −/− embryos, the IX cranial nerve is often fused (arrow) to the X cranial ganglia. (C and D) The same fusion is observed in zf/zf and mz/mz mutants. (E_–_L) Transverse paraffin sections of newborn animals stained with hematoxylin and eosin (dorsal is up). (Scale bar: 200 μm.) The thymus (F_–_H) and parathyroids (J_–_L) are absent in −/−, zf/zf, and mz/mz mutant mice. th, thymus; pt, parathyroid. (M_–_O) Whole-mount in situ hybridization for Pax1 at embryonic d 10.5. Pax1 expression in the third pouch (pp3) is reduced in −/− embryo, but expression in the other pouches is unchanged. zf/zf shows a similar pattern as −/− (cranial is up). (Scale bar: 500 μm.)
Fig. 4.
Novel pharyngeal skeleton morphologies in Hoxa3zf/zf and Hoxa3mz/mz mice, and skeletal phenotype of compound mutants with Hoxd3. (A_–_D) Lateral views of the throat cartilages in cleared newborn skeletal preparations. Anterior is up, dorsal is to the right. Asterisk indicates the lesser horn of hyoid; hy, greater horn of hyoid; thy, thyroid cartilage; crc, cricoid cartilage. (Scale bar: 500 μm.) In Hoxa3null/null (−/−), Hoxa3zf/zf (zf/zf), and Hoxa3mz/mz (mz/mz), the greater horn is malformed and fused to the thyroid cartilage (black arrows in B_–_D). (B) In the null, the lesser horn of the hyoid is greatly reduced or deleted. (C and D) The zf/zf and mz/mz mutants have distinct hyoid morphologies, and are different from WT or null. White arrows show extra cartilage structures in these mutants. (E_–_J) Lateral views of the cervical region in cleared skeleton preparations of the indicated genotypes for Hoxa3 (a3) or Hoxd3 (d3). Anterior is up, dorsal is to the left. Exoccipital (eo) bone, atlas (at), axis (ax), and anterior arch of atlas (arrowhead) are indicated. Note that G and J are similar, whereas I is more similar to H than to F. (Scale bar: 1 mm.)
Fig. 5.
Hoxa3zf allele has null function in NCCs. Hematoxylin and eosin staining of transverse paraffin sections from embryonic d 15.5 embryos (dorsal is up). Genotypes apply to each column; panels in each row are from a comparable anterior–posterior location. In the control embryo, the thymus (th) is located in the chest (A), and the parathyroids (pt) are embedded in the thyroid (F). In embryos with a NCC-specific deletion of mouse Hoxa3 (Hoxa3fx/-;Wnt1cre+/−) the thymic lobes are absent from the normal position (B), and are instead still attached to the pharynx and are ectopic (E). Parathyroids are also ectopic and anterior to the thymus (G). (C, F, and H) Embryos in which only the zf allele is expressed in the NCC (Hoxa3fx_/zf_ ;Wnt1cre+/−) have a phenotype identical to NCC-specific Hoxa3 deletion. (Scale bar: 400 μm.)
Similar articles
- Additional hox clusters in the zebrafish: divergent expression patterns belie equivalent activities of duplicate hoxB5 genes.
Bruce AE, Oates AC, Prince VE, Ho RK. Bruce AE, et al. Evol Dev. 2001 May-Jun;3(3):127-44. doi: 10.1046/j.1525-142x.2001.003003127.x. Evol Dev. 2001. PMID: 11440248 - The Hox code responsible for the patterning of the anterior vertebrae in zebrafish.
Maeno A, Koita R, Nakazawa H, Fujii R, Yamada K, Oikawa S, Tani T, Ishizaka M, Satoh K, Ishizu A, Sugawara T, Adachi U, Kikuchi M, Iwanami N, Matsuda M, Kawamura A. Maeno A, et al. Development. 2024 Jul 15;151(14):dev202854. doi: 10.1242/dev.202854. Epub 2024 Jul 12. Development. 2024. PMID: 38940461 - Spatiotemporal analysis of zebrafish hox gene regulation by Cdx4.
Hayward AG 2nd, Joshi P, Skromne I. Hayward AG 2nd, et al. Dev Dyn. 2015 Dec;244(12):1564-73. doi: 10.1002/dvdy.24343. Epub 2015 Sep 30. Dev Dyn. 2015. PMID: 26335559 - Hox genes in the pharyngeal region: how Hoxa3 controls early embryonic development of the pharyngeal organs.
Gordon J. Gordon J. Int J Dev Biol. 2018;62(11-12):775-783. doi: 10.1387/ijdb.180284jg. Int J Dev Biol. 2018. PMID: 30604847 Review.
Cited by
- Regulatory evolution through divergence of a phosphoswitch in the transcription factor CEBPB.
Lynch VJ, May G, Wagner GP. Lynch VJ, et al. Nature. 2011 Nov 13;480(7377):383-6. doi: 10.1038/nature10595. Nature. 2011. PMID: 22080951 - It's Time to Unite: Diversity and Coordination of Thymic Stromal Cells for T Cell Selection and Organ Integrity.
Muro R, Nitta T. Muro R, et al. Immunol Rev. 2025 Jul;332(1):e70040. doi: 10.1111/imr.70040. Immunol Rev. 2025. PMID: 40464763 Free PMC article. Review. - Mechanisms of thymus organogenesis and morphogenesis.
Gordon J, Manley NR. Gordon J, et al. Development. 2011 Sep;138(18):3865-78. doi: 10.1242/dev.059998. Development. 2011. PMID: 21862553 Free PMC article. Review. - Unraveling the Tangled Skein: The Evolution of Transcriptional Regulatory Networks in Development.
Rebeiz M, Patel NH, Hinman VF. Rebeiz M, et al. Annu Rev Genomics Hum Genet. 2015;16:103-31. doi: 10.1146/annurev-genom-091212-153423. Epub 2015 May 20. Annu Rev Genomics Hum Genet. 2015. PMID: 26079281 Free PMC article. Review. - Evo-devo and accounting for Darwin's endless forms.
Brakefield PM. Brakefield PM. Philos Trans R Soc Lond B Biol Sci. 2011 Jul 27;366(1574):2069-75. doi: 10.1098/rstb.2011.0007. Philos Trans R Soc Lond B Biol Sci. 2011. PMID: 21690125 Free PMC article. Review.
References
- McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. - PubMed
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. - PubMed
- Gellon G, McGinnis W. Shaping animal body plans in development and evolution by modulation of Hox expression patterns. Bioessays. 1998;20:116–125. - PubMed
- Carroll SB. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell. 2008;134:25–36. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases