Evidence for natural antisense transcript-mediated inhibition of microRNA function - PubMed (original) (raw)
Evidence for natural antisense transcript-mediated inhibition of microRNA function
Mohammad Ali Faghihi et al. Genome Biol. 2010.
Abstract
Background: MicroRNAs (miRNAs) have the potential to regulate diverse sets of mRNA targets. In addition, mammalian genomes contain numerous natural antisense transcripts, most of which appear to be non-protein-coding RNAs (ncRNAs). We have recently identified and characterized a highly conserved non-coding antisense transcript for beta-secretase-1 (BACE1), a critical enzyme in Alzheimer's disease pathophysiology. The BACE1-antisense transcript is markedly up-regulated in brain samples from Alzheimer's disease patients and promotes the stability of the (sense) BACE1 transcript.
Results: We report here that BACE1-antisense prevents miRNA-induced repression of BACE1 mRNA by masking the binding site for miR-485-5p. Indeed, miR-485-5p and BACE1-antisense compete for binding within the same region in the open reading frame of the BACE1 mRNA. We observed opposing effects of BACE1-antisense and miR-485-5p on BACE1 protein in vitro and showed that Locked Nucleic Acid-antimiR mediated knockdown of miR-485-5p as well as BACE1-antisense over-expression can prevent the miRNA-induced BACE1 suppression. We found that the expression of BACE1-antisense as well as miR-485-5p are dysregulated in RNA samples from Alzheimer's disease subjects compared to control individuals.
Conclusions: Our data demonstrate an interface between two distinct groups of regulatory RNAs in the computation of BACE1 gene expression. Moreover, bioinformatics analyses revealed a theoretical basis for many other potential interactions between natural antisense transcripts and miRNAs at the binding sites of the latter.
Figures
Figure 1
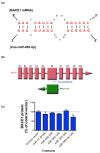
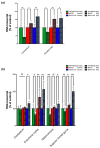
_BACE1_-AS and miR-485-5p competing for the same binding site in BACE1 mRNA. (a) Sequence information of miR-485-5p and its target site in BACE1 mRNA. Binding site in BACE1 mRNA has a strong affinity to the miR-485-5p (free energy = -26.3 using Microinspector; -31.5 using RNA22; -22.8 using miRacle). The predicted target sequence AAGCTGTAGTCAAATCCATCAAGGCAGCCTCC is found within exon 6 of BACE1. (b) The schematic shows the predicted target site for miR-485-5p, the _BACE1_-AS transcript and their relation to BACE1 mRNA. The binding site for miR-485-5p is located in the overlapping region of BACE1 and _BACE1_-AS. BACE1 exons are marked as E1 to E10. Both _BACE1_-AS and miR-485-5p have the potential to bind to exon 6 (E6) of BACE1 mRNA. (c) Over-expression of miR485-5p, but not vectors that over-express miR-17-3p, miR-652, miR-593, or miR-183, nor control empty vector, leads to BACE1 protein reduction by about 30% (**_P_-value < 0.01). Each treatment consists of 24 repeats and error bars represent standard error of means. In this experiment, the miRNA-binding site was not artificially engineered; rather, it is located in its usual place in the open reading frame of the BACE1 transcript. BACE1 protein level was measured by DiscoverRx technology.
Figure 2
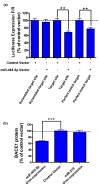
Validation of the miR-485-5p binding site in the BACE1 transcript. (a) The presence of the miR-485-5p target site in the 3' UTR of firefly luciferase is sufficient for depleting luciferase expression by 30%, equally effective as a perfect match positive control. The scrambled target site did not show any effect (**_P_-value < 0.01). This experiment was performed in HEK293T cells and each treatment consisted of 24 biological repeats; error bars represent standard error of means (SEM). (b) Over-expression of miR485-5p, but not control miRNA (miR-219) or empty vector, leads to BACE1 protein reduction by about 30% (***_P_-value < 0.001). Each treatment consists of 32 biological repeats and error bars represent SEM. In this experiment, the miRNA-binding site was not artificially engineered; rather, it is located in its usual place in the open reading frame of the BACE1 transcript. BACE1 protein level was measured by DiscoverRx technology.
Figure 3
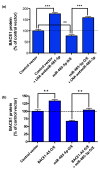
_BACE1_-AS masks the binding site for miR485-5p on BACE1 mRNA. (a) Over-expression (O/E) of miR485-5p significantly reduces BACE1 protein levels in HEK293T C3 cells. BACE1 protein was measured by EFC protein quantification technology (DiscoveRx). LNA-antimir-485-5p, a sequence complementary to the mature miR-485-5p, blocks the effect of miRNA on BACE1 protein expression. LNA-antimiR-485 increases BACE1 protein levels by blocking endogenous miR-485-5p. Each treatment consists of 24 biological repeats and error bars represent standard error of means (SEM; **_P_-value < 0.01 and ***_P_-value < 0.001). (b) Simultaneous over-expression of miR-485-5p and _BACE1_-AS can effectively block the observed BACE1 protein reduction caused by miR-485-5p alone. This indicates that the two ncRNAs can compete for the same binding site on BACE1 mRNA. Each treatment consists of 24 biological repeats and error bars represent SEM (**_P_-value < 0.01).
Figure 4
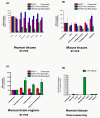
Expression of BACE1, _BACE1_-AS and miR-485-5p in different brain regions. (a) Expression of miR-485-5p, BACE1 and _BACE1_-AS were measured in a commercially available panel of human tissues (n = 1), including brain, liver, heart, skeletal (Sk) muscle, spleen, kidney, testis and lung, by RT-PCR. Whole brain RNA shows a much higher abundance of miR-485-5p (y-axis is log2% of brain). BACE1 mRNA was ubiquitously expressed, with the highest expression in brain. _BACE1_-AS transcript was expressed in all tissues, but relatively higher in brain, heart, skeletal muscle and testis. (b) Expression of miR-485-5p, BACE1 and _BACE1_-AS transcripts were measured in several mouse brain region as well as mouse liver (n = 3). miR-485-5p is readily present in various brain regions, but it is not evenly distributed in all regions tested. BACE1 and _BACE1_-AS transcripts are also highly expressed in all brain regions that are affected by Alzheimer's disease pathologies. (c) Expression of miR-485-5p, BACE1 and _BACE1_-AS transcripts were measured in four human brain regions. RNA originated from cerebellum (18 subjects), entorhinal cortex (8 subjects), hippocampus (12 subjects) and superior frontal gyrus (18 subjects). miR-485-5p was two- to four-fold higher in entorhinal cortex, hippocampus and superior frontal gyrus compared to cerebellum. _BACE1_-AS transcript was expressed two- to three-fold lower in entorhinal cortex, hippocampus and superior frontal gyrus compared to cerebellum. BACE1 mRNA is almost equally distributed in all four regions. (d) The small RNA fraction from two individuals for each of eight tissues (with the exception of the kidney, which had only one sample) was used for high-throughput short read sequencing. After alignment of reads to the human genome, the reads corresponding to miR-485-5p were identified and normalized to the total number of reads. There was significantly higher abundance of miR-485-5p in the orbital gyrus of the brain compared to skeletal muscle, pancreas, lung, heart, liver, spleen and kidney.
Figure 5
Expression of BACE1, _BACE1_-AS and miR-485-5p in Alzheimer's disease-affected individuals. (a) Expression of BACE1, _BACE1_-AS and miR-485-5p transcripts were measured in parietal lobe and cerebellum of five subjects with Alzheimer's disease and five normal elderly individuals. miR-485-5p was down-regulated by 30% in parietal lobe and 60% in cerebellum of Alzheimer's disease patients compared to control individuals. _BACE1_-AS as well as BACE1 transcripts were up-regulated in both cerebellum and parietal lobe (unpaired _t_-test with Welch's correction: ns = not significant; *_P_-value < 0.05; **_P_-value < 0.01; ***_P_-value < 0.001). (b) Expression of BACE1, _BACE1_-AS and miR-485-5p transcripts were measured in four regions of the brain of 35 Alzheimer's disease patients and 35 control individuals. Not all regions were available from all cases; a total of 120 RNA samples from superior frontal gyrus, entorhinal cortex, hippocampus and cerebellum were tested. miR-485-5p was significantly down-regulated in entorhinal cortex and hippocampus of Alzheimer's disease subjects, but not altered in cerebellum nor in superior frontal gyrus. _BACE1_-AS, and to a lesser degree BACE1, transcripts were up-regulated in all four regions. However, the increase in BACE1 mRNA was not statistically significant in cerebellum and hippocampus (unpaired _t_-test with Welch's correction: ns = not significant; *_P_-value < 0.05; **_P_-value < 0.01; ***_P_-value < 0.001)
Figure 6
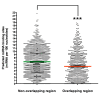
Distribution of miRNA binding sites in sense-antisense RNA transcripts. Predicted miRNA binding sites were counted in 391 human sense-antisense pairs. The total number of predicted miRNAs per 100 nucleotides of overlapping and non-overlapping regions of each sense-antisense pair is depicted. The miRNA binding sites within pairing and non-pairing regions have a median value of 4.78 and 6.49 miRNAs per 100 nucleotides, respectively. On average (mean), each 100 nucleotides of overlapping region have 5.21 miRNA binding sites, while each 100 nucleotides of non-overlapping region have 7.07 miRNA binding sites. The difference seen in miRNA numbers within the sense-antisense pairing regions and non-pairing regions is statistically significant (***_P_-value < 0.0001, Wilcoxon two-sided test).
Similar articles
- Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase.
Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G 3rd, Kenny PJ, Wahlestedt C. Faghihi MA, et al. Nat Med. 2008 Jul;14(7):723-30. doi: 10.1038/nm1784. Epub 2008 Jun 29. Nat Med. 2008. PMID: 18587408 Free PMC article. - BACE1-AS prevents BACE1 mRNA degradation through the sequestration of BACE1-targeting miRNAs.
Zeng T, Ni H, Yu Y, Zhang M, Wu M, Wang Q, Wang L, Xu S, Xu Z, Xu C, Xiong J, Jiang J, Luo Y, Wang Y, Liu H. Zeng T, et al. J Chem Neuroanat. 2019 Jul;98:87-96. doi: 10.1016/j.jchemneu.2019.04.001. Epub 2019 Apr 5. J Chem Neuroanat. 2019. PMID: 30959172 - Research status of the regulation of miRNA on BACE1.
Deng Y, Ding Y, Hou D. Deng Y, et al. Int J Neurosci. 2014 Jul;124(7):474-7. doi: 10.3109/00207454.2013.858249. Epub 2013 Nov 21. Int J Neurosci. 2014. PMID: 24147552 Review. - BACE1 in Alzheimer's disease.
Sathya M, Premkumar P, Karthick C, Moorthi P, Jayachandran KS, Anusuyadevi M. Sathya M, et al. Clin Chim Acta. 2012 Dec 24;414:171-8. doi: 10.1016/j.cca.2012.08.013. Epub 2012 Aug 20. Clin Chim Acta. 2012. PMID: 22926063 Review.
Cited by
- Potential Implications of miRNAs in the Pathogenesis, Diagnosis, and Therapeutics of Alzheimer's Disease.
Wang L, Shui X, Diao Y, Chen D, Zhou Y, Lee TH. Wang L, et al. Int J Mol Sci. 2023 Nov 13;24(22):16259. doi: 10.3390/ijms242216259. Int J Mol Sci. 2023. PMID: 38003448 Free PMC article. Review. - Bidirectional regulation between WDR83 and its natural antisense transcript DHPS in gastric cancer.
Su WY, Li JT, Cui Y, Hong J, Du W, Wang YC, Lin YW, Xiong H, Wang JL, Kong X, Gao QY, Wei LP, Fang JY. Su WY, et al. Cell Res. 2012 Sep;22(9):1374-89. doi: 10.1038/cr.2012.57. Epub 2012 Apr 10. Cell Res. 2012. PMID: 22491477 Free PMC article. - Stabilization of human interferon-α1 mRNA by its antisense RNA.
Kimura T, Jiang S, Nishizawa M, Yoshigai E, Hashimoto I, Nishikawa M, Okumura T, Yamada H. Kimura T, et al. Cell Mol Life Sci. 2013 Apr;70(8):1451-67. doi: 10.1007/s00018-012-1216-x. Epub 2012 Dec 8. Cell Mol Life Sci. 2013. PMID: 23224365 Free PMC article. - Cis-acting noncoding RNAs: friends and foes.
Guil S, Esteller M. Guil S, et al. Nat Struct Mol Biol. 2012 Nov;19(11):1068-75. doi: 10.1038/nsmb.2428. Nat Struct Mol Biol. 2012. PMID: 23132386 Review. - MYC regulates the non-coding transcriptome.
Hart JR, Roberts TC, Weinberg MS, Morris KV, Vogt PK. Hart JR, et al. Oncotarget. 2014 Dec 30;5(24):12543-54. doi: 10.18632/oncotarget.3033. Oncotarget. 2014. PMID: 25587025 Free PMC article.
References
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H. et al.The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources