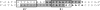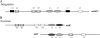CTnDOT integrase interactions with attachment site DNA and control of directionality of the recombination reaction - PubMed (original) (raw)
CTnDOT integrase interactions with attachment site DNA and control of directionality of the recombination reaction
Margaret M Wood et al. J Bacteriol. 2010 Aug.
Abstract
IntDOT is a tyrosine recombinase encoded by the conjugative transposon CTnDOT. The core binding (CB) and catalytic (CAT) domains of IntDOT interact with core-type sites adjacent to the regions of strand exchange, while the N-terminal arm binding (N) domain interacts with arm-type sites distal to the core. Previous footprinting experiments identified five arm-type sites, but how the arm-type sites participate in the integration and excision of CTnDOT was not known. In vitro integration assays with substrates containing arm-type site mutants demonstrated that attDOT sequences containing mutations in the L1 arm-type site or in the R1 and R2 or R1 and R2' arm-type sites were dramatically defective in integration. Substrates containing mutations in the L1 and R1 arm-type sites showed a 10- to 20-fold decrease in detectable in vitro excision, but introduction of multiple arm-type site mutations in attR did not have an effect on the excision frequency. A sixth arm-type site, the R1' site, was also identified and shown to be required for integration and important for efficient excision. These results suggest that intramolecular IntDOT interactions are required for integration, while the actions of accessory factors are more important for excision. Gel shift assays performed in the presence of core- and arm-type site DNAs showed that IntDOT affinity for the attDOT core was enhanced when IntDOT was simultaneously bound to arm-type site DNA.
Figures
FIG. 1.
(A) The CTnDOT core- and arm-type sites. The black boxes indicate positions of arm-type sites. The gray ovals indicate core-type sites. The vertical arrows indicate sites of cleavage. (B) Alignment of CTnDOT arm-type sites. The boldface bases are the regions in each arm-type site that were mutated to a HindIII sequence (5′-AAGCTT-3′), while the underlined region denotes bases that were actually changed as a result of the mutagenesis. Six base pairs were mutated for all of the sites except for the R2 arm-type site, where the sixth-position T was not changed, and the R2′ arm-type site, where the third-position G and fifth-position T were not changed. W represents A or T; D represents A, T, or G. The R1′ arm-type site was a new site identified in this study.
FIG. 2.
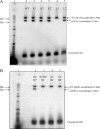
(A) Results of an i n vitro integration competition assay using attDOT sequences containing mutated arm-type site sequences. Radiolabeled attB DNA was incubated with IntDOT, IHF, and two supercoiled plasmids, one containing the wild-type attDOT sequence and one containing a mutation in one of the five arm-type sites. The slower-migrating recombinant in each lane is the larger product resulting from the wild-type internal control, while the faster-migrating recombinant is the product resulting from recombination between the mutant attDOT site and the attB sequence. Lane 1 is a 1-kb ladder. Lane 2 is a control where both the smaller and larger attDOT plasmids contain the wild-type sequence. (B) Results of an in vitro integration competition assay using attDOT sequences containing multiple mutated arm-type sites. Lane 1 is a 1-kb ladder.
FIG. 3.
The R1 region of protection. Bases shaded in gray represent regions protected by IntDOT in DNase I footprinting experiments performed previously, and the boldface type denotes sequence portions previously assigned to R1 (11). The boxes represent bases that were changed in site-directed mutagenesis of the R1′ and R1 arm-type sites.
FIG. 4.
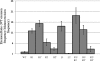
Effects of arm-type site mutations on excision. Excision frequencies were calculated as described in Materials and Methods, and excision ratios were calculated by dividing the wild-type (WT) excision frequency by the mutant excision frequency. Wild-type excision frequencies averaged between 1 and 5%. The values displayed in the figure are the results of at least four independent assays.
FIG. 5.
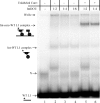
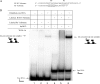
Gel shift analysis of IntDOT complex formation with the L1 arm-type site. Experiments were carried out with labeled wild-type L1 DNA alone (lanes 1 to 4) or labeled wild-type L1 DNA in the presence of unlabeled complementary oligonucleotides containing the attDOT core (lanes 5 and 6). IntDOT was added to the DNA substrates in increasing dilutions, as shown above lanes 2 to 6. An additional band labeled X is visible in all lanes, including the DNA-only lane 1. We believe this band is due to aberrant annealing of the top and bottom oligonucleotides used to make the gel shift substrate. The migration shift of this band in lanes 2 and 5 may be due to interactions of a contaminating DNA binding protein in the partially purified IntDOT preparation. The cartoons diagrammed on the left of the figure indicate possible stoichiometries of the protein-DNA complexes.
FIG. 6.
Gel shift analysis of IntDOT interactions with the attDOT core. Each indicated arm-type site (at approximately 250 nM) was incubated with 40 nM labeled attDOT core and a 1:2 dilution of IntDOT, and the mixtures were subjected to electrophoresis. The cartoons diagrammed on the right of the figure indicate possible stoichiometries of the protein-DNA complexes.
FIG. 7.
(A) DNA sequences containing the R1′ arm-type site or the R1 and R1′ arm-type sites. (B) Gel shift showing IntDOT binding to DNA containing the R1′ or the R1 and R1′ arm-type sites. Approximately 40 nM arm-type site DNA was incubated with IntDOT alone or with IntDOT and unlabeled attDOT core.
FIG. 8.
IntDOT binding to a gel shift substrate containing four arm-type sites. A 120-bp radiolabeled fragment of attDOT was incubated with either 20 or 2 nM IHF and decreasing IntDOT concentrations for 15 min and then electrophoresed on an 8% polyacrylamide gel.
FIG. 9.
(A) Roles of the arm-type sites in integration of CTnDOT. The core-type sites D and D′ are shown as gray ovals. Arm-type sites shown as black boxes are required for integration. Arm-type sites represented by gray boxes were found to be important for intramolecular interactions between IntDOT monomers during integration. Arm-type sites shown in white are not required for integration. (B) Roles of the arm-type sites in excision of CTnDOT. The core-type sites D and D′ are shown as light gray ovals and the core-type sites B and B′ are shown as dark gray ovals. Arm-type sites, represented by gray boxes, stimulate excision, and arm-type sites, shown as white boxes, are not required for excision. (The illustrations not drawn to scale.)
References
- Azaro, M. A., and A. Landy. 2002. Lambda integrase and the lambda Int family, p. 119-148. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, DC.
- Bauer, C. E., S. D. Hesse, R. I. Gumport, and J. F. Gardner. 1986. Mutational analysis of integrase arm-type binding sites of bacteriophage lambda. Integration and excision involve distinct interactions of integrase with arm-type sites. J. Mol. Biol. 192:513-527. - PubMed
- Better, M., S. Wickner, J. Auerbach, and H. Echols. 1983. Role of the Xis protein of bacteriophage lambda in a specific reactive complex at the attR prophage attachment site. Cell 32:161-168. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous