Phenotypic robustness conferred by apparently redundant transcriptional enhancers - PubMed (original) (raw)
. 2010 Jul 22;466(7305):490-3.
doi: 10.1038/nature09158. Epub 2010 May 30.
Affiliations
- PMID: 20512118
- PMCID: PMC2909378
- DOI: 10.1038/nature09158
Phenotypic robustness conferred by apparently redundant transcriptional enhancers
Nicolás Frankel et al. Nature. 2010.
Abstract
Genes include cis-regulatory regions that contain transcriptional enhancers. Recent reports have shown that developmental genes often possess multiple discrete enhancer modules that drive transcription in similar spatio-temporal patterns: primary enhancers located near the basal promoter and secondary, or 'shadow', enhancers located at more remote positions. It has been proposed that the seemingly redundant activity of primary and secondary enhancers contributes to phenotypic robustness. We tested this hypothesis by generating a deficiency that removes two newly discovered enhancers of shavenbaby (svb, a transcript of the ovo locus), a gene encoding a transcription factor that directs development of Drosophila larval trichomes. At optimal temperatures for embryonic development, this deficiency causes minor defects in trichome patterning. In embryos that develop at both low and high extreme temperatures, however, absence of these secondary enhancers leads to extensive loss of trichomes. These temperature-dependent defects can be rescued by a transgene carrying a secondary enhancer driving transcription of the svb cDNA. Finally, removal of one copy of wingless, a gene required for normal trichome patterning, causes a similar loss of trichomes only in flies lacking the secondary enhancers. These results support the hypothesis that secondary enhancers contribute to phenotypic robustness in the face of environmental and genetic variability.
Figures
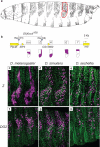
Figure 1. The _svb cis_-regulatory region in D. melanogaster
a, Drawing from the lateral perspective of a D. melanogaster first instar larva. The domain producing quaternary trichomes on the fifth abdominal segment is enclosed in a red outline. b, Diagram of the region upstream of the svb first exon, showing the positions of the five enhancers for this locus: DG2, Z, A, E, and 7. The expression driven by these enhancers in quaternary cells is shown in purple in the diagrams below each enhancer. The piggyBac elements used to generate Df(X)svb108 are shown as blue triangles. c,f, Expression pattern driven by D. melanogaster Z::lacZ (c) and DG2::lacZ (f) in the 5th and 6th abdominal segments of a stage-15 embryo (purple). An anti-Dusky-like antibody was used to stain developing trichomes (green). d,g, Expression pattern driven by D. simulans Z::lacZ (d) and DG2::lacZ (g). e,h, Expression pattern driven by D. sechellia Z::lacZ (e) and DG2::lacZ (h). β-galactosidase protein produced by D. melanogaster Z::lacZ is expressed in the cytoplasm; β-galactosidase from all other constructs is localized to the nucleus.
Figure 2. Effect of Df(X)svb108 on the number of quaternary trichomes
a, The lateral patch (green) and dorsal region (blue) in which trichomes were counted. The green and blue boxes correspond to the regions where the Z and DG2 enhancers are expressed strongly. The primary, secondary, tertiary, and quaternary cell types are indicated with horizontal lines above the photograph. The arrow marks the spiracle that was used to set the lower boundary for the green box. The blue box was positioned directly above the green box. The red box identifies the stout tertiary trichomes, which were excluded from the counts. b, Number of trichomes in the lateral plus dorsal region (blue and green boxes) of the fifth abdominal segment of the larva. Open circles give trichome numbers for each individual (n=10); the black symbols and lines show the mean ±1SD. Embryos from each of the two genotypes (C108 and Df(X)svb108) were reared at three different temperatures: 17°C, 25°C, and 32°C. c, Cuticle images showing the quaternary trichomes in the lateral patch (below) and dorsal region (above) of Df(X)svb108 first-instar larvae that developed at the three different temperatures. The genotype by temperature interaction term of a two-way ANOVA was highly significant (F = 27.57, P<0.0001).
Figure 3. Rescue of the temperature-dependent trichome loss in the lateral patch by a Z::svb transgene
a,b, Trichome number in the lateral patch (a) and dorsal region (b) of the 5th abdominal segment of larvae with the genotypes C108, Df(X)svb108, and Df(X)svb108; Z::svb. Open circles represent trichome numbers for each individual (n=10); the black symbols and lines show the mean ±1SD.
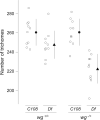
Figure 4. Effect of _Df(X)svb108; wg_-/+ on the number of quaternary trichomes
C108 and Df(X)svb108 embryos that were heterozygous for a null allele of wingless were reared at 25°C. Quaternary trichomes were counted as described in the legend to Fig. 2. A two-way ANOVA reveals a highly significant genotype by temperature interaction (F=7.79, p=0.0084), which is caused by a large reduction in the number of trichomes on _Df(X)svb108; wg_-/+ larvae relative to all other genotypes.
Comment in
- Transcription: Shadow enhancers confer robustness.
Swami M. Swami M. Nat Rev Genet. 2010 Jul;11(7):454. doi: 10.1038/nrg2818. Nat Rev Genet. 2010. PMID: 20548292 No abstract available.
Similar articles
- Genome-wide analyses of Shavenbaby target genes reveals distinct features of enhancer organization.
Menoret D, Santolini M, Fernandes I, Spokony R, Zanet J, Gonzalez I, Latapie Y, Ferrer P, Rouault H, White KP, Besse P, Hakim V, Aerts S, Payre F, Plaza S. Menoret D, et al. Genome Biol. 2013 Aug 23;14(8):R86. doi: 10.1186/gb-2013-14-8-r86. Genome Biol. 2013. PMID: 23972280 Free PMC article. - Multi-enhancer transcriptional hubs confer phenotypic robustness.
Tsai A, Alves MR, Crocker J. Tsai A, et al. Elife. 2019 Jul 11;8:e45325. doi: 10.7554/eLife.45325. Elife. 2019. PMID: 31294690 Free PMC article. - Morphological evolution caused by many subtle-effect substitutions in regulatory DNA.
Frankel N, Erezyilmaz DF, McGregor AP, Wang S, Payre F, Stern DL. Frankel N, et al. Nature. 2011 Jun 29;474(7353):598-603. doi: 10.1038/nature10200. Nature. 2011. PMID: 21720363 Free PMC article. - The structure and evolution of cis-regulatory regions: the shavenbaby story.
Stern DL, Frankel N. Stern DL, et al. Philos Trans R Soc Lond B Biol Sci. 2013 Nov 11;368(1632):20130028. doi: 10.1098/rstb.2013.0028. Print 2013 Dec 19. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 24218640 Free PMC article. Review. - One thousand and one ways of making functionally similar transcriptional enhancers.
Veitia RA. Veitia RA. Bioessays. 2008 Nov;30(11-12):1052-7. doi: 10.1002/bies.20849. Bioessays. 2008. PMID: 18937349 Review.
Cited by
- Regulatory genome annotation of 33 insect species.
Asma H, Tieke E, Deem KD, Rahmat J, Dong T, Huang X, Tomoyasu Y, Halfon MS. Asma H, et al. Elife. 2024 Oct 11;13:RP96738. doi: 10.7554/eLife.96738. Elife. 2024. PMID: 39392676 Free PMC article. - A gene desert required for regulatory control of pleiotropic Shox2 expression and embryonic survival.
Abassah-Oppong S, Zoia M, Mannion BJ, Rouco R, Tissières V, Spurrell CH, Roland V, Darbellay F, Itum A, Gamart J, Festa-Daroux TA, Sullivan CS, Kosicki M, Rodríguez-Carballo E, Fukuda-Yuzawa Y, Hunter RD, Novak CS, Plajzer-Frick I, Tran S, Akiyama JA, Dickel DE, Lopez-Rios J, Barozzi I, Andrey G, Visel A, Pennacchio LA, Cobb J, Osterwalder M. Abassah-Oppong S, et al. Nat Commun. 2024 Oct 10;15(1):8793. doi: 10.1038/s41467-024-53009-7. Nat Commun. 2024. PMID: 39389973 Free PMC article. - Super-silencer perturbation by EZH2 and REST inhibition leads to large loss of chromatin interactions and reduction in cancer growth.
Zhang Y, Chen K, Tang SC, Cai Y, Nambu A, See YX, Fu C, Raju A, Lebeau B, Ling Z, Chan JJ, Tay Y, Mutwil M, Lakshmanan M, Tucker-Kellogg G, Chng WJ, Tenen DG, Osato M, Tergaonkar V, Fullwood MJ. Zhang Y, et al. Nat Struct Mol Biol. 2024 Sep 20. doi: 10.1038/s41594-024-01391-7. Online ahead of print. Nat Struct Mol Biol. 2024. PMID: 39304765 - Evolutionary Innovations in Conserved Regulatory Elements Associate With Developmental Genes in Mammals.
Uebbing S, Kocher AA, Baumgartner M, Ji Y, Bai S, Xing X, Nottoli T, Noonan JP. Uebbing S, et al. Mol Biol Evol. 2024 Oct 4;41(10):msae199. doi: 10.1093/molbev/msae199. Mol Biol Evol. 2024. PMID: 39302728 Free PMC article. - Morphogens in the evolution of size, shape and patterning.
Mosby LS, Bowen AE, Hadjivasiliou Z. Mosby LS, et al. Development. 2024 Sep 15;151(18):dev202412. doi: 10.1242/dev.202412. Epub 2024 Sep 18. Development. 2024. PMID: 39302048 Free PMC article. Review.
References
- Jeong Y, El-Jaick K, Roessler E, Muenke M, Epstein DJ. A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development. 2006;133:761–772. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM063622/GM/NIGMS NIH HHS/United States
- R01 GM063622-09/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- GM063622-06A1/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases