Bayesian integrated modeling of expression data: a case study on RhoG - PubMed (original) (raw)
Bayesian integrated modeling of expression data: a case study on RhoG
Rashi Gupta et al. BMC Bioinformatics. 2010.
Abstract
Background: DNA microarrays provide an efficient method for measuring activity of genes in parallel and even covering all the known transcripts of an organism on a single array. This has to be balanced against that analyzing data emerging from microarrays involves several consecutive steps, and each of them is a potential source of errors. Errors tend to accumulate when moving from the lower level towards the higher level analyses because of the sequential nature. Eliminating such errors does not seem feasible without completely changing the technologies, but one should nevertheless try to meet the goal of being able to realistically assess degree of the uncertainties that are involved when drawing the final conclusions from such analyses.
Results: We present a Bayesian hierarchical model for finding differentially expressed genes between two experimental conditions, proposing an integrated statistical approach where correcting signal saturation, systematic array effects, dye effects, and finding differentially expressed genes, are all modeled jointly. The integration allows all these components, and also the associated errors, to be considered simultaneously. The inference is based on full posterior distribution of gene expression indices and on quantities derived from them rather than on point estimates. The model was applied and tested on two different datasets.
Conclusions: The method presents a way of integrating various steps of microarray analysis into a single joint analysis, and thereby enables extracting information on differential expression in a manner, which properly accounts for various sources of potential error in the process.
Figures
Figure 1
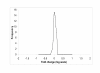
Plot of posterior distribution of D i = T _i_1-T _i_2 for three genes. In the upper panel, posterior distributions of the difference D i = T _i_1-T _i_2 are shown for three genes of dataset-1: a non-differentially expressed gene (left), an up-regulated gene (center), and a down-regulated gene (right). In the lower panel, the corresponding posterior distributions are shown for the latent variable T _i_1 corresponding to the experimental condition (solid line), and for T _i_2 corresponding to the control (dotted line).
Figure 2
Plot of point estimates (posterior means) of log-fold change D i against the overall expression (T _i_1 + T _i_2)/2 for dataset-1. Genes with p _i_+ ≥ 0.99 are plotted with diamonds and those with p _i_- ≥ 0.99 are plotted with triangles. The gene RhoG with p _i_+ = 0.91 is plotted with a red circle.
Figure 3
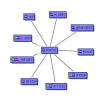
A pictorial representation of the relation of nine genes co-cited with RhoG. The blue boxes (nodes) represent the genes. The "black" edges indicate co-citation of two genes in the PubMed database; the "green" edges indicate a possible regulatory role of JUN and NFKB1 on the expression of RhoG.
Figure 4
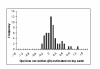
Histograms of point estimates (median of posterior distribution) of D i for GAPDH. These point estimates are of the 56 replicates (on the same array) for a house keeping genes (GAPDH) of dataset-2.
Figure 5
Histogram of point estimates (median of posterior distribution) of β i for GAPDH. These point estimates are of the 56 replicates (on the same array) for a house keeping genes (GAPDH) of dataset-2.
Similar articles
- Bayesian hierarchical error model for analysis of gene expression data.
Cho H, Lee JK. Cho H, et al. Bioinformatics. 2004 Sep 1;20(13):2016-25. doi: 10.1093/bioinformatics/bth192. Epub 2004 Mar 25. Bioinformatics. 2004. PMID: 15044230 - BGX: a fully Bayesian integrated approach to the analysis of Affymetrix GeneChip data.
Hein AM, Richardson S, Causton HC, Ambler GK, Green PJ. Hein AM, et al. Biostatistics. 2005 Jul;6(3):349-73. doi: 10.1093/biostatistics/kxi016. Epub 2005 Apr 14. Biostatistics. 2005. PMID: 15831583 - Bayesian integrated functional analysis of microarray data.
Bhattacharjee M, Pritchard CC, Nelson PS, Arjas E. Bhattacharjee M, et al. Bioinformatics. 2004 Nov 22;20(17):2943-53. doi: 10.1093/bioinformatics/bth338. Epub 2004 Jun 4. Bioinformatics. 2004. PMID: 15180937 - A bayesian hierarchical model for the analysis of Affymetrix arrays.
Tadesse MG, Ibrahim JG. Tadesse MG, et al. Ann N Y Acad Sci. 2004 May;1020:41-8. doi: 10.1196/annals.1310.006. Ann N Y Acad Sci. 2004. PMID: 15208182 Review. - A Bayesian method for analysing spotted microarray data.
Meiklejohn CD, Townsend JP. Meiklejohn CD, et al. Brief Bioinform. 2005 Dec;6(4):318-30. doi: 10.1093/bib/6.4.318. Brief Bioinform. 2005. PMID: 16420731 Review.
Cited by
- Segmentation and intensity estimation for microarray images with saturated pixels.
Yang Y, Stafford P, Kim Y. Yang Y, et al. BMC Bioinformatics. 2011 Nov 30;12:462. doi: 10.1186/1471-2105-12-462. BMC Bioinformatics. 2011. PMID: 22129216 Free PMC article. - BASiCS: Bayesian Analysis of Single-Cell Sequencing Data.
Vallejos CA, Marioni JC, Richardson S. Vallejos CA, et al. PLoS Comput Biol. 2015 Jun 24;11(6):e1004333. doi: 10.1371/journal.pcbi.1004333. eCollection 2015 Jun. PLoS Comput Biol. 2015. PMID: 26107944 Free PMC article.
References
- Dudoit S, Yang YH, Luu P, Speed TP. In: Microarrays:Optical Technolologies and Informatics, Vol. 4266 of Proceedings of SPIE. Bittner ML, Chen Y, Dorsel AN, Dougherty ER, editor. Vol. 2001. Normalization for cDNA microarray data; pp. 141–152.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources