Phylogenetic evidence for lateral gene transfer in the intestine of marine iguanas - PubMed (original) (raw)
Phylogenetic evidence for lateral gene transfer in the intestine of marine iguanas
David M Nelson et al. PLoS One. 2010.
Abstract
Background: Lateral gene transfer (LGT) appears to promote genotypic and phenotypic variation in microbial communities in a range of environments, including the mammalian intestine. However, the extent and mechanisms of LGT in intestinal microbial communities of non-mammalian hosts remains poorly understood.
Methodology/principal findings: We sequenced two fosmid inserts obtained from a genomic DNA library derived from an agar-degrading enrichment culture of marine iguana fecal material. The inserts harbored 16S rRNA genes that place the organism from which they originated within Clostridium cluster IV, a well documented group that habitats the mammalian intestinal tract. However, sequence analysis indicates that 52% of the protein-coding genes on the fosmids have top BLASTX hits to bacterial species that are not members of Clostridium cluster IV, and phylogenetic analysis suggests that at least 10 of 44 coding genes on the fosmids may have been transferred from Clostridium cluster XIVa to cluster IV. The fosmids encoded four transposase-encoding genes and an integrase-encoding gene, suggesting their involvement in LGT. In addition, several coding genes likely involved in sugar transport were probably acquired through LGT.
Conclusion: Our phylogenetic evidence suggests that LGT may be common among phylogenetically distinct members of the phylum Firmicutes inhabiting the intestinal tract of marine iguanas.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
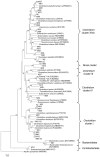
Figure 1. Phylogenetic relationships of 16S rRNA gene sequences among the fosmids, clone library, and Clostridium clusters.
Clone library sequences start with “M.” The numbers in parentheses following some of the marine iguana sequences indicate the number of times that a particular sequence was obtained. Only representatives of the major Clostridium clusters, and limited representatives of the Bacteroidetes and Coriobacteriales, are shown. The tree was inferred using the neighbor joining approach. The numbers at the nodes represent bootstrap values. The bar represents 0.02 substitutions per nucleotide position. The outgroup is Aquifex pyrophilus.
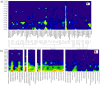
Figure 2. BLASTP result distribution across fosmids 7–14 and 7–25.
a) The X-axis indicates genera with at least 10 BLASTP hits throughout the ORFeome of the analyzed fosmids. Using a previously published approach the organism distribution on a genus level was identified for each coding gene, e-values were grouped into ranges, and threshold levels were defined for minimum overall frequency. Genera are phylogenetically sorted. The Y-axis indicates respective e-value ranges. The frequency of hits for each genus in each e-value range (log scale) is shown by color coding and corresponding values are indicated in the figure. All BLASTP hits per genus per ORF were accepted. b) Same as a), except that custom databases of species from phylogenetically defined Clostridium clusters were used.
Similar articles
- Metagenomic-based study of the phylogenetic and functional gene diversity in Galápagos land and marine iguanas.
Hong PY, Mao Y, Ortiz-Kofoed S, Shah R, Cann I, Mackie RI. Hong PY, et al. Microb Ecol. 2015 Feb;69(2):444-56. doi: 10.1007/s00248-014-0547-6. Epub 2014 Dec 19. Microb Ecol. 2015. PMID: 25524569 - Lateral gene transfer and phylogenetic assignment of environmental fosmid clones.
Nesbø CL, Boucher Y, Dlutek M, Doolittle WF. Nesbø CL, et al. Environ Microbiol. 2005 Dec;7(12):2011-26. doi: 10.1111/j.1462-2920.2005.00918.x. Environ Microbiol. 2005. PMID: 16309397 - The complete mitochondrial genomes of the Galápagos iguanas, Amblyrhynchus cristatus and Conolophus subcristatus.
MacLeod A, Irisarri I, Vences M, Steinfartz S. MacLeod A, et al. Mitochondrial DNA A DNA Mapp Seq Anal. 2016 Sep;27(5):3699-700. doi: 10.3109/19401736.2015.1079863. Epub 2015 Sep 10. Mitochondrial DNA A DNA Mapp Seq Anal. 2016. PMID: 26357924 - Trends and barriers to lateral gene transfer in prokaryotes.
Popa O, Dagan T. Popa O, et al. Curr Opin Microbiol. 2011 Oct;14(5):615-23. doi: 10.1016/j.mib.2011.07.027. Epub 2011 Aug 17. Curr Opin Microbiol. 2011. PMID: 21856213 Review. - Detecting lateral genetic transfer : a phylogenetic approach.
Beiko RG, Ragan MA. Beiko RG, et al. Methods Mol Biol. 2008;452:457-69. doi: 10.1007/978-1-60327-159-2_21. Methods Mol Biol. 2008. PMID: 18566777 Review.
Cited by
- Defining reference sequences for Nocardia species by similarity and clustering analyses of 16S rRNA gene sequence data.
Helal M, Kong F, Chen SC, Bain M, Christen R, Sintchenko V. Helal M, et al. PLoS One. 2011;6(6):e19517. doi: 10.1371/journal.pone.0019517. Epub 2011 Jun 8. PLoS One. 2011. PMID: 21687706 Free PMC article. - Diets Alter the Gut Microbiome of Crocodile Lizards.
Jiang HY, Ma JE, Li J, Zhang XJ, Li LM, He N, Liu HY, Luo SY, Wu ZJ, Han RC, Chen JP. Jiang HY, et al. Front Microbiol. 2017 Oct 25;8:2073. doi: 10.3389/fmicb.2017.02073. eCollection 2017. Front Microbiol. 2017. PMID: 29118742 Free PMC article. - Captivity affects diversity, abundance, and functional pathways of gut microbiota in the northern grass lizard Takydromus septentrionalis.
Zhou J, Zhao YT, Dai YY, Jiang YJ, Lin LH, Li H, Li P, Qu YF, Ji X. Zhou J, et al. Microbiologyopen. 2020 Sep;9(9):e1095. doi: 10.1002/mbo3.1095. Epub 2020 Jul 14. Microbiologyopen. 2020. PMID: 32666685 Free PMC article. - Phylogenetic analysis of the fecal microbial community in herbivorous land and marine iguanas of the Galápagos Islands using 16S rRNA-based pyrosequencing.
Hong PY, Wheeler E, Cann IK, Mackie RI. Hong PY, et al. ISME J. 2011 Sep;5(9):1461-70. doi: 10.1038/ismej.2011.33. Epub 2011 Mar 31. ISME J. 2011. PMID: 21451584 Free PMC article. - A phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria.
Meehan CJ, Beiko RG. Meehan CJ, et al. Genome Biol Evol. 2014 Mar;6(3):703-13. doi: 10.1093/gbe/evu050. Genome Biol Evol. 2014. PMID: 24625961 Free PMC article.
References
- Browne J, Neve M, editors. The Voyage of the Beagle: Charles Darwin's Journal of Researches. 1989. 448. Penguin Classics (abridged edition)
- Wikelski M, Thom C. Marine iguanas shrink to survive El Niño-Changes in bone metabolism enable these adult lizards to reversibly alter their length. Nature. 2000;403:37–38. - PubMed
- Throckmorton GS. Oral food-processing in 2 herbivorous lizards, Iguana iguana (Iguanidae) and Uromastix aegyptius (Agamidae). Journal of Morphology. 1976;148:363–390. - PubMed
- Mackie RI, Nelson DM, Wheeler E, Wikelski M, Cann IKO. Fermentative digestion in herbivorous lizards: bacterial population analysis in the intestinal tract of free-living land (Conolophus pallidus) and marine iguanas (Amblyrynchus cristatus) on the Galápagos archipelago. In: Morris S, Vosloo A, editors. 4th Comparative Physiology and Biochemistry Meeting in Africa: Mara 2008 Molecules to Migration: The Pressures of Life. Bologna, Italy: Medimond Publishing Co; 2008. pp. 193–202.
- Mackie RI, White BA, Isaacson RE, editors. New York: Springer-Verlag; 1997. Gastrointestinal Microbiology: Gastrointestinal Microbes and Host Interactions.676
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources