Transcriptional profiling of Clostridium difficile and Caco-2 cells during infection - PubMed (original) (raw)
Transcriptional profiling of Clostridium difficile and Caco-2 cells during infection
Tavan Janvilisri et al. J Infect Dis. 2010.
Abstract
Clostridium difficile is well recognized as the most common infectious cause of nosocomial diarrhea. The incidence and severity of C. difficile infection (CDI) is increasing worldwide. Here, we evaluated simultaneously the transcriptional changes in the human colorectal epithelial Caco-2 cells and in C. difficile after infection. A total of 271 transcripts in Caco-2 cells and 207 transcripts in C. difficile were significantly differentially expressed at 1 time point during CDI. We used the gene ontology annotations and protein-protein network interactions to underline a framework of target molecules that could potentially play a key role during CDI. These genes included those associated with cellular metabolism, transcription, transport, cell communication, and signal transduction. Our data identified certain key factors that have previously been reported to be involved in CDI, as well as novel determinants that may participate in a complex mechanism underlying the host response to infection, bacterial adaptation, and pathogenesis.
Conflict of interest statement
All authors listed in the title page do not have a commercial or other association that might pose a conflict of interest.
Figures
Figure 1
Infection of Caco-2 cells with C. difficile. (A) Quantification of Caco-2 cell viability at different times post-infection. All assays were conducted in triplicate and repeated independently three times. Cell viability is as expressed as the percentage of survival of the control wells. Results are expressed as means ± 1 standard deviation for the replicate experiments, and the Student t test was used for statistical analysis of the data. Significant difference from the control (p < 0.05) is indicated by an asterisk. Caco-2 cells under anaerobic conditions at different time points without the infection were also evaluated but they were not significantly different from the control cells. (B) Photomicrographs of infected Caco-2 cells with C. difficile at 30, 60 and 120 min post-infection.
Figure 2
Transcriptional dialogue between Caco-2 cells and C. difficile during infection. (A) The number of up- or down-regulated genes after 30, 60, and 120 min p.i., as compared to the expression levels at the time of infection. (B, C) Hierarchical clustering analysis of differentially expressed genes in Caco-2 cells and C. difficile during infection. Genes identified to be significantly differentially expressed at 30, 60 or 120 min in Caco-2 or C. difficile cells p.i. relative to in vitro growth. Genes significantly different with _p_-value < 0.05 after the infection were pooled and used to create heatmaps for (B) Caco-2 cells, and (C) C. difficile. Genes are ordered in rows, conditions as columns. Red color indicates genes induced post-infection vs. prior to infection (fold change); green color denotes repression.
Figure 3
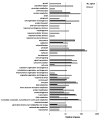
Functional annotation of genes in Caco-2 cells and C. difficile, which are differentially expressed between infected and uninfected conditions. All differentially expressed genes were annotated using generic GO-slim for biological process.
Figure 4
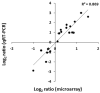
Validation of microarray data by qRT-PCR. Gene expression changes in infected versus uninfected cells measured by microarray analysis or qRT-PCR are compared. Data are plotted as log2 ratios of microarray data (x-axis) compared to those of qRT-PCR (y-axis).
Similar articles
- Identification of the role of toxin B in the virulence of Clostridioides difficile based on integrated bioinformatics analyses.
Gao Y, Gao W, Cheng J, Ma L, Su J. Gao Y, et al. Int Microbiol. 2020 Nov;23(4):575-587. doi: 10.1007/s10123-020-00128-y. Epub 2020 May 9. Int Microbiol. 2020. PMID: 32388701 - Clostridium difficile Toxin B causes epithelial cell necrosis through an autoprocessing-independent mechanism.
Chumbler NM, Farrow MA, Lapierre LA, Franklin JL, Haslam DB, Goldenring JR, Lacy DB. Chumbler NM, et al. PLoS Pathog. 2012;8(12):e1003072. doi: 10.1371/journal.ppat.1003072. Epub 2012 Dec 6. PLoS Pathog. 2012. PMID: 23236283 Free PMC article. - Hypervirulent Clostridium difficile strains in hospitalized patients, Canada.
Mulvey MR, Boyd DA, Gravel D, Hutchinson J, Kelly S, McGeer A, Moore D, Simor A, Suh KN, Taylor G, Weese JS, Miller M; Canadian Nosocomial Infection Surveillance Program. Mulvey MR, et al. Emerg Infect Dis. 2010 Apr;16(4):678-81. doi: 10.3201/eid1604.091152. Emerg Infect Dis. 2010. PMID: 20350386 Free PMC article. - Microarray identification of Clostridium difficile core components and divergent regions associated with host origin.
Janvilisri T, Scaria J, Thompson AD, Nicholson A, Limbago BM, Arroyo LG, Songer JG, Gröhn YT, Chang YF. Janvilisri T, et al. J Bacteriol. 2009 Jun;191(12):3881-91. doi: 10.1128/JB.00222-09. Epub 2009 Apr 17. J Bacteriol. 2009. PMID: 19376880 Free PMC article. - Clostridium difficile infection: new developments in epidemiology and pathogenesis.
Rupnik M, Wilcox MH, Gerding DN. Rupnik M, et al. Nat Rev Microbiol. 2009 Jul;7(7):526-36. doi: 10.1038/nrmicro2164. Nat Rev Microbiol. 2009. PMID: 19528959 Review.
Cited by
- The complete catalog of antimicrobial resistance secondary active transporters in Clostridioides difficile: evolution and drug resistance perspective.
Chanket W, Pipatthana M, Sangphukieo A, Harnvoravongchai P, Chankhamhaengdecha S, Janvilisri T, Phanchana M. Chanket W, et al. Comput Struct Biotechnol J. 2024 May 21;23:2358-2374. doi: 10.1016/j.csbj.2024.05.027. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 38873647 Free PMC article. - Investigation of metabolic crosstalk between host and pathogenic Clostridioides difficile via multiomics approaches.
Kwon JE, Jo SH, Song WS, Lee JS, Jeon HJ, Park JH, Kim YR, Baek JH, Kim MG, Kwon SY, Kim JS, Yang YH, Kim YG. Kwon JE, et al. Front Bioeng Biotechnol. 2022 Sep 2;10:971739. doi: 10.3389/fbioe.2022.971739. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36118584 Free PMC article. - Plakoglobin and High-Mobility Group Box 1 Mediate Intestinal Epithelial Cell Apoptosis Induced by Clostridioides difficile TcdB.
Li Y, Xu W, Ren Y, Cheung HC, Huang P, Kaur G, Kuo CJ, McDonough SP, Fubini SL, Lipkin SM, Deng X, Chang YF, Huang L. Li Y, et al. mBio. 2022 Oct 26;13(5):e0184922. doi: 10.1128/mbio.01849-22. Epub 2022 Aug 31. mBio. 2022. PMID: 36043787 Free PMC article. - FliW and CsrA Govern Flagellin (FliC) Synthesis and Play Pleiotropic Roles in Virulence and Physiology of Clostridioides difficile R20291.
Zhu D, Wang S, Sun X. Zhu D, et al. Front Microbiol. 2021 Oct 5;12:735616. doi: 10.3389/fmicb.2021.735616. eCollection 2021. Front Microbiol. 2021. PMID: 34675903 Free PMC article. - Predictive regulatory and metabolic network models for systems analysis of Clostridioides difficile.
Arrieta-Ortiz ML, Immanuel SRC, Turkarslan S, Wu WJ, Girinathan BP, Worley JN, DiBenedetto N, Soutourina O, Peltier J, Dupuy B, Bry L, Baliga NS. Arrieta-Ortiz ML, et al. Cell Host Microbe. 2021 Nov 10;29(11):1709-1723.e5. doi: 10.1016/j.chom.2021.09.008. Epub 2021 Oct 11. Cell Host Microbe. 2021. PMID: 34637780 Free PMC article.
References
- Kelly CP, LaMont JT. Clostridium difficile infection. Annu Rev Med. 1998;49:375–90. - PubMed
- Arroyo LG, Kruth SA, Willey BM, Staempfli HR, Low DE, Weese JS. PCR ribotyping of Clostridium difficile isolates originating from human and animal sources. J Med Microbiol. 2005;54:163–6. - PubMed
- McDonald LC, Killgore GE, Thompson A, et al. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med. 2005;353:2433–41. - PubMed
- Chang JY, Antonopoulos DA, Kalra A, et al. Decreased diversity of the fecal Microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis. 2008;197:435–8. - PubMed
- Jank T, Giesemann T, Aktories K. Rho-glycosylating Clostridium difficile toxins A and B: new insights into structure and function. Glycobiology. 2007;17:15R–22R. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical