ABA receptors: the START of a new paradigm in phytohormone signalling - PubMed (original) (raw)
Review
. 2010 Jul;61(12):3199-210.
doi: 10.1093/jxb/erq151. Epub 2010 Jun 3.
Affiliations
- PMID: 20522527
- PMCID: PMC3107536
- DOI: 10.1093/jxb/erq151
Review
ABA receptors: the START of a new paradigm in phytohormone signalling
John P Klingler et al. J Exp Bot. 2010 Jul.
Abstract
The phytohormone abscisic acid (ABA) plays a central role in plant development and in plant adaptation to both biotic and abiotic stressors. In recent years, knowledge of ABA metabolism and signal transduction has advanced rapidly to provide detailed glimpses of the hormone's activities at the molecular level. Despite this progress, many gaps in understanding have remained, particularly at the early stages of ABA perception by the plant cell. The search for an ABA receptor protein has produced multiple candidates, including GCR2, GTG1, and GTG2, and CHLH. In addition to these candidates, in 2009 several research groups converged on a novel family of Arabidopsis proteins that bind ABA, and thereby interact directly with a class of protein phosphatases that are well known as critical players in ABA signal transduction. The PYR/PYL/RCAR receptor family is homologous to the Bet v 1-fold and START domain proteins. It consists of 14 members, nearly all of which appear capable of participating in an ABA receptor-signal complex that responds to the hormone by activating the transcription of ABA-responsive genes. Evidence is provided here that PYR/PYL/RCAR receptors can also drive the phosphorylation of the slow anion channel SLAC1 to provide a fast and timely response to the ABA signal. Crystallographic studies have vividly shown the mechanics of ABA binding to PYR/PYL/RCAR receptors, presenting a model that bears some resemblance to the binding of gibberellins to GID1 receptors. Since this ABA receptor family is highly conserved in crop species, its discovery is likely to usher a new wave of progress in the elucidation and manipulation of plant stress responses in agricultural settings.
Figures
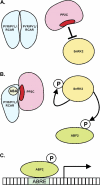
Fig. 1.
Schematic view of the ABA signal–receptor complex, including a PYR/PYL/RCAR homodimer (A) and hormone-bound protomer (B); a PP2C phosphatase, with the active site indicated in dark red; a SnRK2 kinase; and the ABF2 transcription factor. In the absence of ABA (A), the receptor forms a homodimer, while the PP2C inhibits both autophosphorylation of the SnRK2 and phosphorylation of ABF2. In the presence of ABA (B), a receptor protomer engulfs the hormone within a pocket, allowing the receptor to bind the PP2C and cover the phosphatase active site. This permits the autophosphorylation of the SnRK2 and phosphorylation of its ABF2 substrate. In its phosphorylated, active state (C), ABF2 binds to an ABA-responsive element (ABRE) in the promoter of ABA-responsive genes, activating transcription.
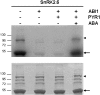
Fig. 2.
Regulation of SLAC1 phosphorylation status by the ABA-dependent PYR1, ABI1, SnRK2.6 signalling cascade. GST-SLAC1 N-terminal fragment (SLAC N, Met1 to Phe188) was incubated in the presence of [γ-32P]ATP with MBP-SnRK2.6 pretreated without (–) or with (+) GST-ABI1, His-PYR1, and 100 μM (±)-ABA. Bands of SLAC N fragment and MBP–SnRK2.6 are indicated by an arrow and an arrowhead, respectively. Coomassie-stained SDS-PAGE (bottom) and autoradiogram of the gel (top) are shown. Recombinant proteins and reaction conditions were as described previously (Fujii et al., 2009; Park et al., 2009). When SLAC N was incubated with SnRK2.6 not treated with GST-ABI1, both SnRK2.6 autophosphorylation and SLAC N phosphorylation bands were visualized (first lane). SnRK2.6 pre-treated with GST-ABI1 was unable to phosphorylate SLAC N (second lane). In the absence of ABA, the addition of His-PYR1 to the pretreatment of SnRK2.6 with GST-ABI could not restore SnRK2.6 phosphorylation activity on SLAC N (third lane). However, when ABA was added to the pretreatment reaction, SLAC N phosphorylation was recovered (fourth lane).
Fig. 3.
Amino acid sequence alignment of the Arabidopsis PYR1 protein with the most similar homologues in five cultivated species. The alignment was performed with the CLUSTALW2 program (
http://www.ebi.ac.uk/Tools/clustalw2/index.html
) using the default settings. Asterisks indicate residues in contact with ABA hormone, according to the PYL2 crystal structure of Melcher et al. (2009). The locations of the gate and latch domains are indicated.
Similar articles
- PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis.
Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, Lee S, Caddell DF, Cutler SR, Chory J, Yates JR, Schroeder JI. Nishimura N, et al. Plant J. 2010 Jan;61(2):290-9. doi: 10.1111/j.1365-313X.2009.04054.x. Epub 2009 Oct 26. Plant J. 2010. PMID: 19874541 Free PMC article. - The ABA signal transduction mechanism in commercial crops: learning from Arabidopsis.
Ben-Ari G. Ben-Ari G. Plant Cell Rep. 2012 Aug;31(8):1357-69. doi: 10.1007/s00299-012-1292-2. Epub 2012 Jun 4. Plant Cell Rep. 2012. PMID: 22660953 Review. - C2-domain abscisic acid-related proteins mediate the interaction of PYR/PYL/RCAR abscisic acid receptors with the plasma membrane and regulate abscisic acid sensitivity in Arabidopsis.
Rodriguez L, Gonzalez-Guzman M, Diaz M, Rodrigues A, Izquierdo-Garcia AC, Peirats-Llobet M, Fernandez MA, Antoni R, Fernandez D, Marquez JA, Mulet JM, Albert A, Rodriguez PL. Rodriguez L, et al. Plant Cell. 2014 Dec;26(12):4802-20. doi: 10.1105/tpc.114.129973. Epub 2014 Dec 2. Plant Cell. 2014. PMID: 25465408 Free PMC article. - A Novel Chemical Inhibitor of ABA Signaling Targets All ABA Receptors.
Ye Y, Zhou L, Liu X, Liu H, Li D, Cao M, Chen H, Xu L, Zhu JK, Zhao Y. Ye Y, et al. Plant Physiol. 2017 Apr;173(4):2356-2369. doi: 10.1104/pp.16.01862. Epub 2017 Feb 13. Plant Physiol. 2017. PMID: 28193765 Free PMC article. - Structural insights into PYR/PYL/RCAR ABA receptors and PP2Cs.
Santiago J, Dupeux F, Betz K, Antoni R, Gonzalez-Guzman M, Rodriguez L, Márquez JA, Rodriguez PL. Santiago J, et al. Plant Sci. 2012 Jan;182:3-11. doi: 10.1016/j.plantsci.2010.11.014. Epub 2010 Dec 7. Plant Sci. 2012. PMID: 22118610 Review.
Cited by
- Molecular mechanisms in the activation of abscisic acid receptor PYR1.
Dorosh L, Kharenko OA, Rajagopalan N, Loewen MC, Stepanova M. Dorosh L, et al. PLoS Comput Biol. 2013;9(6):e1003114. doi: 10.1371/journal.pcbi.1003114. Epub 2013 Jun 27. PLoS Comput Biol. 2013. PMID: 23825939 Free PMC article. - Structural basis and functions of abscisic acid receptors PYLs.
Zhang XL, Jiang L, Xin Q, Liu Y, Tan JX, Chen ZZ. Zhang XL, et al. Front Plant Sci. 2015 Feb 19;6:88. doi: 10.3389/fpls.2015.00088. eCollection 2015. Front Plant Sci. 2015. PMID: 25745428 Free PMC article. Review. - Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions.
Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. Hubbard KE, et al. Genes Dev. 2010 Aug 15;24(16):1695-708. doi: 10.1101/gad.1953910. Genes Dev. 2010. PMID: 20713515 Free PMC article. Review. - Populus trichocarpa clade A PP2C protein phosphatases: their stress-induced expression patterns, interactions in core abscisic acid signaling, and potential for regulation of growth and development.
Rigoulot SB, Petzold HE, Williams SP, Brunner AM, Beers EP. Rigoulot SB, et al. Plant Mol Biol. 2019 Jun;100(3):303-317. doi: 10.1007/s11103-019-00861-7. Epub 2019 Apr 3. Plant Mol Biol. 2019. PMID: 30945147 - Developing a model of plant hormone interactions.
Wang YH, Irving HR. Wang YH, et al. Plant Signal Behav. 2011 Apr;6(4):494-500. doi: 10.4161/psb.6.4.14558. Epub 2011 Apr 1. Plant Signal Behav. 2011. PMID: 21406974 Free PMC article. Review.
References
- Cao YJ, Wei Q, Liao Y, Song HL, Li X, Xiang CB, Kuai BK. Ectopic overexpression of AtHDG11 in tall fescue resulted in enhanced tolerance to drought and salt stress. Plant Cell Reports. 2009;28:579–588. - PubMed
- Chini A, Fonseca S, Fernandez G, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases