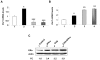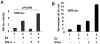Mutually positive regulatory feedback loop between interferons and estrogen receptor-alpha in mice: implications for sex bias in autoimmunity - PubMed (original) (raw)
Mutually positive regulatory feedback loop between interferons and estrogen receptor-alpha in mice: implications for sex bias in autoimmunity
Ravichandran Panchanathan et al. PLoS One. 2010.
Abstract
Background: Systemic lupus erythematosus (SLE), an autoimmune disease, predominantly affects women of childbearing age. Moreover, increased serum levels of interferon-alpha (IFN-alpha) are associated with the disease. Although, the female sex hormone estrogen (E2) is implicated in sex bias in SLE through up-regulation of IFN-gamma expression, the molecular mechanisms remain unknown. Here we report that activation of IFN (alpha or gamma)-signaling in immune cells up-regulates expression of estrogen receptor-alpha (ERalpha; encoded by the Esr1 gene) and stimulates expression of target genes.
Methodology/principal findings: We found that treatment of mouse splenic cells and mouse cell lines with IFN (alpha or gamma) increased steady-state levels of ERalpha mRNA and protein. The increase in the ERalpha mRNA levels was primarily due to the transcriptional mechanisms and it was dependent upon the activation of signal transducer and activator of transcription-1 (STAT1) factor by IFN. Moreover, the IFN-treatment of cells also stimulated transcription of a reporter gene, expression of which was driven by the promoter region of the murine Esr1 gene. Notably, splenic cells from pre-autoimmune lupus-prone (NZB x NZW)F(1) female mice had relatively higher steady-state levels of mRNAs encoded by the IFN and ERalpha-responsive genes as compared to the age-matched males.
Conclusions/significance: Our observations identify a novel mutually positive regulatory feedback loop between IFNs and ERalpha in immune cells in mice and support the idea that activation of this regulatory loop contributes to sex bias in SLE.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. IFN-treatment increases steady-state levels of ERα mRNA and protein in C57BL/6 splenic cells.
(A) Total RNA was isolated from control (column 1), IFN-α (column 2), or IFN-γ (column 3) treated total splenic cells that were prepared from female (age ∼8 weeks) C57BL/6 mice. The RNA was analyzed for steady-sate levels of Esr1 mRNA by quantitative real-time PCR. The ratio of the Esr1 mRNA to β2-microglobulin mRNA was calculated in units (one unit being the ratio of Esr1 mRNA to β2-microglobulin mRNA). Results are mean values of triplicate experiments and error bars represent standard deviation (* p<0.05). (B) Total RNA was isolated from control (column 1 and 3) or IFN-α (column 2 and 4) treated splenic cells that were prepared from either male (age ∼8 weeks) or age-matched female C57BL/6 mice. The RNA was analyzed by quantitative real-time PCR for the steady-sate levels of Esr1 mRNA as described in (A). Results are mean values of triplicate experiments and error bars represent standard deviation (* p<0.05; ** p<0.005). (C) Total protein extracts were prepared from control (lanes 1 and 3) or IFN-α (lanes 2 and 4) treated splenic cells that were isolated from either male (age ∼8 weeks) or age-matched female C57BL/6 mice. The total cell extracts were analyzed by immunoblotting using antibodies specific to the indicated proteins. Fold change in ERα protein levels is indicated below the Figure. (D) Total RNA was isolated from purified splenic T cells (column 1) or B cells (column 2) isolated from female (age ∼8 weeks) C57BL/6 mice. The RNA was analyzed by quantitative real-time PCR for steady-sate levels of Esr1 mRNA. Results are mean values of triplicate experiments and error bars represent the standard deviation (** p<0.005).
Figure 2. IFN-treatment increases steady-state levels of ERα mRNA and protein in splenic cells from lupus-prone (NZB × NZW) F1 mice.
(A) Total RNA isolated from control (lanes 1 and 3) or IFN-α (lanes 2 and 4) treated total splenic cells that were isolated from either male (age ∼8 weeks) or age-matched female (NZB × NZW) F1 mice. The total RNA was analyzed for steady-state levels of Esr1 mRNA by semi-quantitative RT-PCR. Fold change in Esr1 mRNA levels is indicated below the Figure. (B) Total RNA isolated from control (columns 1 and 3) or IFN-α (columns 2 and 4) treated total splenic cells that were isolated from either male (age ∼8 weeks) or age-matched female (NZB × NZW) F1 mice. The total RNA was analyzed for the steady-state levels of Esr1 mRNA by quantitative real-time PCR. Results are mean values of triplicate experiments and error bars represent standard deviation (* p<0.05; NS, not significant). (C) Total protein extracts were prepared from control (lanes 1 and 3) or IFN-α (lanes 2 and 4) treated splenic cells that were isolated from either male (age ∼8 weeks) or age-matched female (NZB × NZW)F1 mice. The total cell extracts were analyzed by immunoblotting using antibodies specific to the indicated proteins. Fold change in ERα protein levels is indicated below the Figure. (D and E) Total protein extracts were prepared from control (lanes 1 and 3) or IFN-α (lanes 2 and 4) treated purified splenic B cells that were isolated from either male (age ∼8 weeks) or age-matched female (NZB × NZW) F1 mice. The total cell extracts were analyzed by immunoblotting using antibodies specific to the indicated proteins. Fold change in ERα protein levels is indicated below the Figures.
Figure 3. IFN-treatment increases steady-state levels of ERα mRNA and protein in mouse breast cancer cell line WT276.
(A) Total protein extracts were prepared from control (lane 1), increasing concentrations (1,000 or 2000 u/ml) of IFN-α (lanes 2 and 3, respectively) or IFN-γ (5 or 10 ng/ml; lanes 4 and 5, respectively) treated WT276 cells. As a negative control, we also included extracts from AKR-2B cells. The extracts were analyzed by immunoblotting using the antibodies specific to the indicated proteins. Fold change in ERα protein levels is indicated below the Figure. (B) Total RNA was isolated from control (lane 1), increasing concentrations of IFN-α (lanes 2 and 3) or IFN-γ (lanes 4 and 5) treated WT276 cells. As a positive control, we also included RNA from splenic cells. The total RNA was analyzed for steady-state levels of Esr1 mRNA by semi-quantitative RT-PCR. Fold change in ERα mRNA levels is indicated below the Figure. (C) Total RNA was isolated from control (column 1), increasing concentrations of IFN-α (columns 2 and 3) or IFN-γ (columns 4 and 5) treated WT276 cells. The total RNA was analyzed for steady-state levels of Esr1 mRNA by quantitative real-time PCR. Results are mean values of triplicate experiments and error bars represent standard deviation (* p<0.05).
Figure 4. Interferon-signaling increases ERα mRNA levels primarily by Transcriptional mechanism.
(A) Total RNA was isolated from control (column 1), IFN-α (column 2), actinomycin D (column 3), or both IFN-α and actinomycin D (column 4) treated WT276 cells. The RNA was analyzed by quantitative real-time PCR for steady-sate levels of Esr1 mRNA. Results are mean values of triplicate experiments and error bars represent standard deviation (* p<0.05; *** p<0.0005). (B) Total RNA was isolated from control (column 1), IFN-α (column 2), cycloheximide (column 3), or both IFN-α and cycloheximide (column 4) treated WT276 cells. The RNA was analyzed by quantitative real-time PCR for steady-sate levels of Esr1 mRNA. Results are mean values of triplicate experiments and error bars represent standard deviation (* p<0.05; ** p<0.005). (C) Total cell extracts were prepared from control (lane 1), IFN-α (lane 2), cycloheximide (lane 3), or both IFN-α and cycloheximide (lane 4) treated WT276 cells. The cell extracts were analyzed by immunoblotting using antibodies specific to the indicated proteins. Fold change in ERα protein levels is indicated below the Figure.
Figure 5. Expression of the Esr1 gene is dependent on activation of STAT1.
(A)
Top panel:
Schematic presentation of the 5′-regulatory region of the murine Esr1 gene (the NCBI accession # for the sequence: NT_039490.7) and potential _cis_-elements that are predicted to render the gene responsive to the IFN treatment. The regulatory sequence for the gene is derived from the C57BL/6J strain of mice. The regulatory region includes three potential ISREs for binding of activated STAT1 (ISGF3) transcription factor.
Bottom panel:
Sub-confluent cultures of WT276 cells in a 6-well plate were transfected with ERα promoter-β-galactosidase plasmid (2 µg) along with pRL-TK (0.2 µg) plasmid using the FuGene 6 transfection agent. 24 h after transfections, cells were either left untreated treated (column1), treated with IFN-α (column 2), or IFN-γ (column 3). 40–45 h after transfections, cells were lysed and the lysates were processed for estimation of protein followed by β-galactosidase activity assays. (B) Total RNA was isolated from wilt-type (columns 1 and 3) or STAT1-null (columns 2 and 4) total splenic cells that were prepared from male or age-matched female (age ∼8 weeks) mice. The RNA was analyzed by quantitative real-time PCR for steady-sate levels of Esr1 mRNA. (C) Total cell extracts were prepared from wilt-type (lanes 1 and 3) or STAT1-null (lanes 2 and 4) total splenic cells that were prepared from male or age-matched female (age ∼8 weeks) mice. The RNA was analyzed by quantitative real-time PCR for steady-sate levels of Esr1 mRNA. Fold change in ERα protein levels is indicated below the Figure. (D) Total cell isolated from C57BL/6 were either left untreated (lane 1) or treated with fludarabine (lane 2) for 24 h. Total cell extracts were prepared and analyzed by immunoblotting using antibodies specific to the indicated proteins. Fold change in ERα protein levels is indicated below the Figure. (E) Total cell isolated from C57BL/6 were either left untreated (lane 1) or treated with fludarabine (lane 2) for 24 h. Total RNA was prepared and steady-state levels of Esr1 mRNA were analyzed by quantitative real-time PCR. Results are mean values of triplicate experiments and error bars represent standard deviation (** p<0.005).
Figure 6. The IFN and E2-signaling cooperate to activate transcription of genes.
(A) Sub-confluent cultures of WT276 cells in a 6-well plate were transfected with ERE-luc-reporter plasmid (2 µg) along with pRL-TK (0.2 µg) plasmid using FuGENE 6 transfection reagent. 24 h after transfections, cells were either left untreated (column 1), treated with E2 (column 2), IFN-α (column 3) or E2 and IFN-α (column 4). 40–45 h after transfections, cells were processed for dual luciferase activity. Result are mean values of triplicate experiments and error bars represent standard deviation (p value is 0.006). (B) Sub-confluent cultures of WT276 cells in a 6-well plate were transfected with ISRE-luc-reporter plasmid (2 µg) along with pRL-TK (0.2 µg) plasmid using FuGENE 6 transfection reagent. 24 h after transfections, cells were either left untreated (column 1), treated with E2 (column 2), IFN-α (column 3) or E2 and IFN-α (column 4). 40–45 h after transfections, cells were processed for dual luciferase activity.
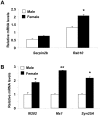
Figure 7. Sex bias in the expression of IFN and E2-responsive genes.
(A and B) Total RNA isolated from pre-autoimmune (age ∼8 weeks) male or age-matched female (NZB × NZW) F1 mice was analyzed for steady-state levels of the indicated known estrogen-responsive (A) and IFN-responsive (B) genes by quantitative real-time PCR. Results are mean values of triplicate experiments and error bars represent standard deviation (* p<0.05; ** p<0.005).
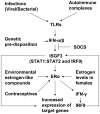
Figure 8. Cooperation between the IFN and E2-signaling in sex bias in SLE in mice.
Increased levels of type I IFNs up-regulate expression of ERα. Activation of ERα by the female sex hormone estrogen leads to up-regulation of IFN-γ and IFN-γ-inducible IRF9. Increased levels of the IRF9 potentiate ISGF3-mediated transcription of IFN-inducible genes, which mediate the immunomodulatory functions of the IFNs.
Similar articles
- Murine BAFF expression is up-regulated by estrogen and interferons: implications for sex bias in the development of autoimmunity.
Panchanathan R, Choubey D. Panchanathan R, et al. Mol Immunol. 2013 Jan;53(1-2):15-23. doi: 10.1016/j.molimm.2012.06.013. Epub 2012 Jul 10. Mol Immunol. 2013. PMID: 22784990 Free PMC article. - Female and male sex hormones differentially regulate expression of Ifi202, an interferon-inducible lupus susceptibility gene within the Nba2 interval.
Panchanathan R, Shen H, Bupp MG, Gould KA, Choubey D. Panchanathan R, et al. J Immunol. 2009 Dec 1;183(11):7031-8. doi: 10.4049/jimmunol.0802665. Epub 2009 Nov 4. J Immunol. 2009. PMID: 19890043 Free PMC article. - Expression of murine Unc93b1 is up-regulated by interferon and estrogen signaling: implications for sex bias in the development of autoimmunity.
Panchanathan R, Liu H, Choubey D. Panchanathan R, et al. Int Immunol. 2013 Sep;25(9):521-9. doi: 10.1093/intimm/dxt015. Epub 2013 Jun 1. Int Immunol. 2013. PMID: 23728775 Free PMC article. - Signaling Pathways of Type I and Type III Interferons and Targeted Therapies in Systemic Lupus Erythematosus.
Chyuan IT, Tzeng HT, Chen JY. Chyuan IT, et al. Cells. 2019 Aug 23;8(9):963. doi: 10.3390/cells8090963. Cells. 2019. PMID: 31450787 Free PMC article. Review. - A Positive Feedback Amplifier Circuit That Regulates Interferon (IFN)-Stimulated Gene Expression and Controls Type I and Type II IFN Responses.
Michalska A, Blaszczyk K, Wesoly J, Bluyssen HAR. Michalska A, et al. Front Immunol. 2018 May 28;9:1135. doi: 10.3389/fimmu.2018.01135. eCollection 2018. Front Immunol. 2018. PMID: 29892288 Free PMC article. Review.
Cited by
- Interferons in autoimmune and inflammatory diseases: regulation and roles.
Choubey D, Moudgil KD. Choubey D, et al. J Interferon Cytokine Res. 2011 Dec;31(12):857-65. doi: 10.1089/jir.2011.0101. J Interferon Cytokine Res. 2011. PMID: 22149411 Free PMC article. Review. - Sex Hormones in Acquired Immunity and Autoimmune Disease.
Moulton VR. Moulton VR. Front Immunol. 2018 Oct 4;9:2279. doi: 10.3389/fimmu.2018.02279. eCollection 2018. Front Immunol. 2018. PMID: 30337927 Free PMC article. Review. - Host responses to the pathogen Mycobacterium avium subsp. paratuberculosis and beneficial microbes exhibit host sex specificity.
Karunasena E, McMahon KW, Chang D, Brashears MM. Karunasena E, et al. Appl Environ Microbiol. 2014 Aug;80(15):4481-90. doi: 10.1128/AEM.01229-14. Appl Environ Microbiol. 2014. PMID: 24814797 Free PMC article. - Tetrabromobisphenol A activates the hepatic interferon pathway in rats.
Dunnick JK, Morgan DL, Elmore SA, Gerrish K, Pandiri A, Ton TV, Shockley KR, Merrick BA. Dunnick JK, et al. Toxicol Lett. 2017 Jan 15;266:32-41. doi: 10.1016/j.toxlet.2016.11.019. Epub 2016 Nov 30. Toxicol Lett. 2017. PMID: 27914987 Free PMC article. - Lupus-prone B6.Nba2 male and female mice display anti-DWEYS reactivity and a neuropsychiatric phenotype.
Browne K, Zhang E, Sullivan JK, Evonuk KS, DeSilva TM, Jorgensen TN. Browne K, et al. Brain Behav Immun. 2021 May;94:175-184. doi: 10.1016/j.bbi.2021.02.010. Epub 2021 Feb 17. Brain Behav Immun. 2021. PMID: 33607233 Free PMC article.
References
- Kotzin BL. Systemic lupus erythematosus. Cell. 1996;85:303–306. - PubMed
- Whitacre CC. Sex differences in autoimmune disease. Nat Immunol 2: 777- 2001;780 - PubMed
- Rider V, Abdou NI. Gender differences in autoimmunity: molecular basis for estrogen effects in systemic lupus erythematosus. Int Immunopharmacol. 2001;1:1009–1024. - PubMed
- Cohen-Solal JF, Jeganathan V, Grimaldi CM, Peeva E, Diamond B. Sex hormones and SLE: influencing the fate of auto reactive B cells. Curr Top Microbiol Immunol. 2006;305:67–88. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous