Mass spectrometric analysis, automated identification and complete annotation of O-linked glycopeptides - PubMed (original) (raw)
Mass spectrometric analysis, automated identification and complete annotation of O-linked glycopeptides
Zsuzsa Darula et al. Eur J Mass Spectrom (Chichester). 2010.
Abstract
Complex mixtures containing O-linked glycopeptides bearing SA(1-0)GalGalNAc structures, or single GalNAc units were subjected to collision-induced dissociation (CID) and electron transfer dissociation (ETD) analysis on a linear ion trap-Orbitrap mass spectrometer and the resulting data was analyzed using the Protein Prospector software. An overview of the structural information provided by the different fragmentation techniques, as well as their limitations, is presented. We illustrate the importance of the complementary information in the mass spectrometry survey scans as well as the different tandem mass spectrometry techniques. We also present some unique features offered by Protein Prospector that are advantageous in glycopeptide analysis: (i) considering a modification that will produce a neutral loss, without "labeling" the original modification site; (ii) merging CID and ETD search results; (iii) permitting the comparison of different modification site-assignments. Although these data were obtained from secreted glycopeptides, the observations and conclusions are also valid for the intracellular regulatory O-GlcNAc modification.
Figures
Figure 1
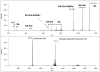
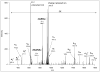
CID and ETD spectra of a 3+ precursor ion at m/z 848.39 (MS spectrum is presented in Figure 4). The upper panel shows the CID spectrum acquired in the linear ion trap; the spectrum is dominated by fragments that consist of the intact glycopeptide minus sugar units, annotated “–SA” (the glycopeptide minus sialic acid), “-SA-Gal”, etc. There are also non-reducing end oxonium ions (“SA”, “SA-Gal”, etc.). The lower panel shows the ETD spectrum. This spectrum contains no useful information, probably because the low charge-density of the precursor ion prevented efficient ETD fragmentation.
Figure 2
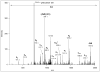
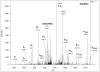
ETD spectrum of m/z 733.0369(3+), corresponding to AAT(GalNAcGalSA)LSTLAGQPLLER. This spectrum provides sufficient information for confident sequence identification as well as site assignment. Sialic acid loss from charge-reduced versions of the precursor ion was also detected. Fragments labeled with asterisk indicate hydrogen transfer, i.e. z+1 ions.
Figure 3
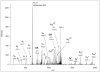
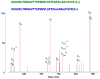
This ETD spectrum of m/z 642.03 (4+) was manually deciphered as representing the Na-adduct of TEELQQQNTAPT(GalNAcGalSA)NSPTK. CID and ETD data for the same glycopeptide from a protonated ion of a lower charge state are shown in Figure 1. Its MS spectrum is presented in Figure 4, where the upper panel clearly illustrates that at the higher charge state the solely protonated ion was practically non-existent. Asterisk-labeled fragments retained the Na-ion.
Figure 4
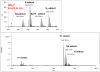
MS survey scan showing the multiply charged precursor ion clusters for the glycopeptide presented in Figures 1 and 3. The MS scan also illustrates that the distribution of protonated ions and Na- and K-adducts can be significantly different at different charge states. Thus, sometimes only metal-ion adducts will be subjected to MS/MS analysis from a cluster.
Figure 5
ETD data from precursor ion 632.6347 (3+), in the linear ion trap. A partially deglycosylated mixture was analyzed, and the ALRPSPT(GalNAc)S(GalNAc)PPSENH glycopeptide was identified from these data. Fragments labeled with asterisk indicate hydrogen transfer, i.e. c-1· and z+1 ions.
Figure 6
CID data of m/z 1157.0476, acquired in the linear ion trap from a partially deglycosylated mixture. Fragment ions labeled with G retained the sugar unit. The structure is DVSASTTVLPDDVT(GalNAc)AYPVG bearing an additional GalNAc unit on any of the 4 other hydroxy amino acids.
Figure 7
Fragment assignment of a glycopeptide CID using MS-Product from SearchCompare output of ProteinProspector. The green y fragment assignments indicate gas-phase deglycosylation, while the glycosylated y5 and y6 ions (in blue) show that there is sufficient information to pinpoint the modification site.
Similar articles
- Software for automated interpretation of mass spectrometry data from glycans and glycopeptides.
Woodin CL, Maxon M, Desaire H. Woodin CL, et al. Analyst. 2013 May 21;138(10):2793-803. doi: 10.1039/c2an36042j. Analyst. 2013. PMID: 23293784 Free PMC article. Review. - Selective detection and site-analysis of O-GlcNAc-modified glycopeptides by beta-elimination and tandem electrospray mass spectrometry.
Greis KD, Hayes BK, Comer FI, Kirk M, Barnes S, Lowary TL, Hart GW. Greis KD, et al. Anal Biochem. 1996 Feb 1;234(1):38-49. doi: 10.1006/abio.1996.0047. Anal Biochem. 1996. PMID: 8742080 - Use of CID/ETD mass spectrometry to analyze glycopeptides.
Mechref Y. Mechref Y. Curr Protoc Protein Sci. 2012 Apr;Chapter 12:12.11.1-12.11.11. doi: 10.1002/0471140864.ps1211s68. Curr Protoc Protein Sci. 2012. PMID: 22470127 Free PMC article. - Glycoproteomics based on tandem mass spectrometry of glycopeptides.
Wuhrer M, Catalina MI, Deelder AM, Hokke CH. Wuhrer M, et al. J Chromatogr B Analyt Technol Biomed Life Sci. 2007 Apr 15;849(1-2):115-28. doi: 10.1016/j.jchromb.2006.09.041. Epub 2006 Oct 17. J Chromatogr B Analyt Technol Biomed Life Sci. 2007. PMID: 17049937 Review.
Cited by
- A calibration routine for efficient ETD in large-scale proteomics.
Rose CM, Rush MJ, Riley NM, Merrill AE, Kwiecien NW, Holden DD, Mullen C, Westphall MS, Coon JJ. Rose CM, et al. J Am Soc Mass Spectrom. 2015 Nov;26(11):1848-57. doi: 10.1007/s13361-015-1183-1. Epub 2015 Jun 26. J Am Soc Mass Spectrom. 2015. PMID: 26111518 Free PMC article. - Precursor ion survival energies of protonated N-glycopeptides and their weak dependencies on high mannose N-glycan composition in collision-induced dissociation.
Aboufazeli F , Dodds ED . Aboufazeli F , et al. Analyst. 2018 Sep 10;143(18):4459-4468. doi: 10.1039/c8an00830b. Analyst. 2018. PMID: 30151520 Free PMC article. - Software for automated interpretation of mass spectrometry data from glycans and glycopeptides.
Woodin CL, Maxon M, Desaire H. Woodin CL, et al. Analyst. 2013 May 21;138(10):2793-803. doi: 10.1039/c2an36042j. Analyst. 2013. PMID: 23293784 Free PMC article. Review. - N- and O-glycosylation in the murine synaptosome.
Trinidad JC, Schoepfer R, Burlingame AL, Medzihradszky KF. Trinidad JC, et al. Mol Cell Proteomics. 2013 Dec;12(12):3474-88. doi: 10.1074/mcp.M113.030007. Epub 2013 Jul 1. Mol Cell Proteomics. 2013. PMID: 23816992 Free PMC article. - Parallel Determination of Polypeptide and Oligosaccharide Connectivities by Energy-Resolved Collison-Induced Dissociation of Protonated O-Glycopeptides Derived from Nonspecific Proteolysis.
Kelly MI, Dodds ED. Kelly MI, et al. J Am Soc Mass Spectrom. 2020 Mar 4;31(3):624-632. doi: 10.1021/jasms.9b00065. Epub 2020 Feb 21. J Am Soc Mass Spectrom. 2020. PMID: 32126781 Free PMC article.
References
- Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. Cold Spring Harbor Laboratory Press; New York: 2009. - PubMed
- Hofsteenge J, Muller DR, de Beer T, Loffler A, Richter WJ, Vliegenthart JF. New type of linkage between a carbohydrate and a protein: C-glycosylation of a specific tryptophan residue in human Rnase. Biochemistry. 1994;33:13524. - PubMed
- Medzihradszky KF, Gillece-Castro BL, Settineri CA, Townsend RR, Masiarz FR, Burlingame AL. Structure Determination of O-Linked Glycopeptides by Tandem Mass-Spectrometry. Biomed Environ Mass Spectrom. 1990;19:777. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P41 GM103481/GM/NIGMS NIH HHS/United States
- RR001614/RR/NCRR NIH HHS/United States
- S10 RR019934-01/RR/NCRR NIH HHS/United States
- RR019934/RR/NCRR NIH HHS/United States
- S10 RR019934/RR/NCRR NIH HHS/United States
- P41 RR001614-27/RR/NCRR NIH HHS/United States
- P41 RR001614/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources