Functional impact of global rare copy number variation in autism spectrum disorders - PubMed (original) (raw)
. 2010 Jul 15;466(7304):368-72.
doi: 10.1038/nature09146. Epub 2010 Jun 9.
Alistair T Pagnamenta, Lambertus Klei, Richard Anney, Daniele Merico, Regina Regan, Judith Conroy, Tiago R Magalhaes, Catarina Correia, Brett S Abrahams, Joana Almeida, Elena Bacchelli, Gary D Bader, Anthony J Bailey, Gillian Baird, Agatino Battaglia, Tom Berney, Nadia Bolshakova, Sven Bölte, Patrick F Bolton, Thomas Bourgeron, Sean Brennan, Jessica Brian, Susan E Bryson, Andrew R Carson, Guillermo Casallo, Jillian Casey, Brian H Y Chung, Lynne Cochrane, Christina Corsello, Emily L Crawford, Andrew Crossett, Cheryl Cytrynbaum, Geraldine Dawson, Maretha de Jonge, Richard Delorme, Irene Drmic, Eftichia Duketis, Frederico Duque, Annette Estes, Penny Farrar, Bridget A Fernandez, Susan E Folstein, Eric Fombonne, Christine M Freitag, John Gilbert, Christopher Gillberg, Joseph T Glessner, Jeremy Goldberg, Andrew Green, Jonathan Green, Stephen J Guter, Hakon Hakonarson, Elizabeth A Heron, Matthew Hill, Richard Holt, Jennifer L Howe, Gillian Hughes, Vanessa Hus, Roberta Igliozzi, Cecilia Kim, Sabine M Klauck, Alexander Kolevzon, Olena Korvatska, Vlad Kustanovich, Clara M Lajonchere, Janine A Lamb, Magdalena Laskawiec, Marion Leboyer, Ann Le Couteur, Bennett L Leventhal, Anath C Lionel, Xiao-Qing Liu, Catherine Lord, Linda Lotspeich, Sabata C Lund, Elena Maestrini, William Mahoney, Carine Mantoulan, Christian R Marshall, Helen McConachie, Christopher J McDougle, Jane McGrath, William M McMahon, Alison Merikangas, Ohsuke Migita, Nancy J Minshew, Ghazala K Mirza, Jeff Munson, Stanley F Nelson, Carolyn Noakes, Abdul Noor, Gudrun Nygren, Guiomar Oliveira, Katerina Papanikolaou, Jeremy R Parr, Barbara Parrini, Tara Paton, Andrew Pickles, Marion Pilorge, Joseph Piven, Chris P Ponting, David J Posey, Annemarie Poustka, Fritz Poustka, Aparna Prasad, Jiannis Ragoussis, Katy Renshaw, Jessica Rickaby, Wendy Roberts, Kathryn Roeder, Bernadette Roge, Michael L Rutter, Laura J Bierut, John P Rice, Jeff Salt, Katherine Sansom, Daisuke Sato, Ricardo Segurado, Ana F Sequeira, Lili Senman, Naisha Shah, Val C Sheffield, Latha Soorya, Inês Sousa, Olaf Stein, Nuala Sykes, Vera Stoppioni, Christina Strawbridge, Raffaella Tancredi, Katherine Tansey, Bhooma Thiruvahindrapduram, Ann P Thompson, Susanne Thomson, Ana Tryfon, John Tsiantis, Herman Van Engeland, John B Vincent, Fred Volkmar, Simon Wallace, Kai Wang, Zhouzhi Wang, Thomas H Wassink, Caleb Webber, Rosanna Weksberg, Kirsty Wing, Kerstin Wittemeyer, Shawn Wood, Jing Wu, Brian L Yaspan, Danielle Zurawiecki, Lonnie Zwaigenbaum, Joseph D Buxbaum, Rita M Cantor, Edwin H Cook, Hilary Coon, Michael L Cuccaro, Bernie Devlin, Sean Ennis, Louise Gallagher, Daniel H Geschwind, Michael Gill, Jonathan L Haines, Joachim Hallmayer, Judith Miller, Anthony P Monaco, John I Nurnberger Jr, Andrew D Paterson, Margaret A Pericak-Vance, Gerard D Schellenberg, Peter Szatmari, Astrid M Vicente, Veronica J Vieland, Ellen M Wijsman, Stephen W Scherer, James S Sutcliffe, Catalina Betancur
Affiliations
- PMID: 20531469
- PMCID: PMC3021798
- DOI: 10.1038/nature09146
Functional impact of global rare copy number variation in autism spectrum disorders
Dalila Pinto et al. Nature. 2010.
Abstract
The autism spectrum disorders (ASDs) are a group of conditions characterized by impairments in reciprocal social interaction and communication, and the presence of restricted and repetitive behaviours. Individuals with an ASD vary greatly in cognitive development, which can range from above average to intellectual disability. Although ASDs are known to be highly heritable ( approximately 90%), the underlying genetic determinants are still largely unknown. Here we analysed the genome-wide characteristics of rare (<1% frequency) copy number variation in ASD using dense genotyping arrays. When comparing 996 ASD individuals of European ancestry to 1,287 matched controls, cases were found to carry a higher global burden of rare, genic copy number variants (CNVs) (1.19 fold, P = 0.012), especially so for loci previously implicated in either ASD and/or intellectual disability (1.69 fold, P = 3.4 x 10(-4)). Among the CNVs there were numerous de novo and inherited events, sometimes in combination in a given family, implicating many novel ASD genes such as SHANK2, SYNGAP1, DLGAP2 and the X-linked DDX53-PTCHD1 locus. We also discovered an enrichment of CNVs disrupting functional gene sets involved in cellular proliferation, projection and motility, and GTPase/Ras signalling. Our results reveal many new genetic and functional targets in ASD that may lead to final connected pathways.
Figures
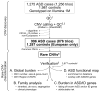
Figure 1. CNV discovery and characterization
Comprehensive procedures were used to identify the rare CNV dataset (boxed). Dashed arrows indicate CNVs not included in downstream analyses. SNP and intensity quality control (QC) with ancestry estimation. QC for CNV calls. Pilot validation experiments using quantitative-PCR were used to evaluate the false discovery-rate. Rare CNVs in samples of EA ancestry were defined as 30 kb in size and present in the total sample set at a frequency <1%. 70/996 (17%) of ASD cases were analyzed on different lower-resolution arrays in previous studies,,. All CNVs were computationally verified and at least 40% of case-CNVs were also experimentally validated by qPCR and/or independent Agilent or other SNP microarrays. 3,677 additional EA controls were used to test specific loci from the primary burden analyses. Additional details are in the Methods Summary and Supplementary Information
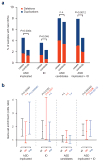
Figure 2. CNV burden in known ASD and/or ID genes
a, Proportion of samples with CNVs overlapping genes and loci known to be associated in ASD with or without ID or ID only, as well as published candidate genes and loci for ASD (Supplementary Table 9). To select for CNVs with maximal impact, they needed to intersect genes, and overlap the target loci by ≥50% of their length. Fisher’s exact test _P_-values for significant differences (_P_≤0.05, one tailed) are shown. b, enrichment analysis for genes overlapped by rare CNVs in cases compared to controls for the three gene-sets in panel a, relative to the whole genome. Odds ratio (OR) and 95% confidence intervals are given for each gene set. Empirical _P_-values for gene-set enrichment are indicated above each OR. All _P_-values <0.1 are listed.
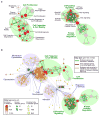
Figure 3. A functional map of ASD
Enrichment results were mapped into a network of gene-sets (nodes) related by mutual overlap (edges), where the color (red, blue, or yellow) indicates the class of gene-set. Node size is proportional to the total number of genes in each set and edge thickness represents the number of overlapping genes between sets. a, Gene-sets enriched for deletions are shown (red) with enrichment significance (FDR q-value) represented as a node color gradient. Groups of functionally related gene-sets are circled and labeled (groups, solid line; sub-groups, dashed line). b, An expanded enrichment map shows the relationship between gene-sets enriched in deletions (panel a) and sets of known ASD/ID genes. Node color hue represents the class of gene-set (i.e. enriched in deletions, red; known disease genes (ie. ASD and/or ID genes), blue; enriched only in disease genes, yellow). Edge color represents the overlap between gene-sets enriched in deletions (green), from disease genes to enriched sets (blue), and between sets enriched in deletions and in disease genes or between disease gene-sets only (orange). The major functional groups are highlighted by filled circles (enriched in deletions, green; enriched in ASD/ID, blue).
Similar articles
- A discovery resource of rare copy number variations in individuals with autism spectrum disorder.
Prasad A, Merico D, Thiruvahindrapuram B, Wei J, Lionel AC, Sato D, Rickaby J, Lu C, Szatmari P, Roberts W, Fernandez BA, Marshall CR, Hatchwell E, Eis PS, Scherer SW. Prasad A, et al. G3 (Bethesda). 2012 Dec;2(12):1665-85. doi: 10.1534/g3.112.004689. Epub 2012 Dec 1. G3 (Bethesda). 2012. PMID: 23275889 Free PMC article. - Rare Inherited and De Novo CNVs Reveal Complex Contributions to ASD Risk in Multiplex Families.
Leppa VM, Kravitz SN, Martin CL, Andrieux J, Le Caignec C, Martin-Coignard D, DyBuncio C, Sanders SJ, Lowe JK, Cantor RM, Geschwind DH. Leppa VM, et al. Am J Hum Genet. 2016 Sep 1;99(3):540-554. doi: 10.1016/j.ajhg.2016.06.036. Epub 2016 Aug 25. Am J Hum Genet. 2016. PMID: 27569545 Free PMC article. - Genetic architecture in autism spectrum disorder.
Devlin B, Scherer SW. Devlin B, et al. Curr Opin Genet Dev. 2012 Jun;22(3):229-37. doi: 10.1016/j.gde.2012.03.002. Epub 2012 Mar 29. Curr Opin Genet Dev. 2012. PMID: 22463983 Review. - Rare copy number variants in tourette syndrome disrupt genes in histaminergic pathways and overlap with autism.
Fernandez TV, Sanders SJ, Yurkiewicz IR, Ercan-Sencicek AG, Kim YS, Fishman DO, Raubeson MJ, Song Y, Yasuno K, Ho WS, Bilguvar K, Glessner J, Chu SH, Leckman JF, King RA, Gilbert DL, Heiman GA, Tischfield JA, Hoekstra PJ, Devlin B, Hakonarson H, Mane SM, Günel M, State MW. Fernandez TV, et al. Biol Psychiatry. 2012 Mar 1;71(5):392-402. doi: 10.1016/j.biopsych.2011.09.034. Epub 2011 Dec 14. Biol Psychiatry. 2012. PMID: 22169095 Free PMC article. - [Autism spectrum disorder and genes for synaptic proteins].
Shishido E. Shishido E. Brain Nerve. 2012 Jan;64(1):65-70. Brain Nerve. 2012. PMID: 22223503 Review. Japanese.
Cited by
- Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease.
Cooper DN, Krawczak M, Polychronakos C, Tyler-Smith C, Kehrer-Sawatzki H. Cooper DN, et al. Hum Genet. 2013 Oct;132(10):1077-130. doi: 10.1007/s00439-013-1331-2. Epub 2013 Jul 3. Hum Genet. 2013. PMID: 23820649 Free PMC article. Review. - Genotype to phenotype relationships in autism spectrum disorders.
Chang J, Gilman SR, Chiang AH, Sanders SJ, Vitkup D. Chang J, et al. Nat Neurosci. 2015 Feb;18(2):191-8. doi: 10.1038/nn.3907. Epub 2014 Dec 22. Nat Neurosci. 2015. PMID: 25531569 Free PMC article. - The disruption of Celf6, a gene identified by translational profiling of serotonergic neurons, results in autism-related behaviors.
Dougherty JD, Maloney SE, Wozniak DF, Rieger MA, Sonnenblick L, Coppola G, Mahieu NG, Zhang J, Cai J, Patti GJ, Abrahams BS, Geschwind DH, Heintz N. Dougherty JD, et al. J Neurosci. 2013 Feb 13;33(7):2732-53. doi: 10.1523/JNEUROSCI.4762-12.2013. J Neurosci. 2013. PMID: 23407934 Free PMC article. - A genome-wide association study of autism using the Simons Simplex Collection: Does reducing phenotypic heterogeneity in autism increase genetic homogeneity?
Chaste P, Klei L, Sanders SJ, Hus V, Murtha MT, Lowe JK, Willsey AJ, Moreno-De-Luca D, Yu TW, Fombonne E, Geschwind D, Grice DE, Ledbetter DH, Mane SM, Martin DM, Morrow EM, Walsh CA, Sutcliffe JS, Lese Martin C, Beaudet AL, Lord C, State MW, Cook EH Jr, Devlin B. Chaste P, et al. Biol Psychiatry. 2015 May 1;77(9):775-84. doi: 10.1016/j.biopsych.2014.09.017. Epub 2014 Sep 30. Biol Psychiatry. 2015. PMID: 25534755 Free PMC article. - A genome-wide copy number variant study of suicidal behavior.
Gross JA, Bureau A, Croteau J, Galfalvy H, Oquendo MA, Haghighi F, Mérette C, Giegling I, Hodgkinson C, Goldman D, Rujescu D, Mann JJ, Turecki G. Gross JA, et al. PLoS One. 2015 May 26;10(5):e0128369. doi: 10.1371/journal.pone.0128369. eCollection 2015. PLoS One. 2015. PMID: 26010658 Free PMC article.
References
- Veenstra-Vanderweele J, Christian SL, Cook EH., Jr Autism as a paradigmatic complex genetic disorder. Annu Rev Genomics Hum Genet. 2004;5:379–405. - PubMed
- Chakrabarti S, Fombonne E. Pervasive developmental disorders in preschool children: confirmation of high prevalence. Am J Psychiatry. 2005;162 (6):1133–1141. - PubMed
- Bailey A, et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25 (1):63–77. - PubMed
Publication types
MeSH terms
Grants and funding
- P01 NS026630/NS/NINDS NIH HHS/United States
- U19 HD035469-06/HD/NICHD NIH HHS/United States
- R01 MH080647-11/MH/NIMH NIH HHS/United States
- MH06359/MH/NIMH NIH HHS/United States
- R01 DA019963-01A2/DA/NIDA NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 DA019963-03/DA/NIDA NIH HHS/United States
- P50 HD055748-02/HD/NICHD NIH HHS/United States
- MH57881/MH/NIMH NIH HHS/United States
- P50 HD055782/HD/NICHD NIH HHS/United States
- R01 MH061009-05/MH/NIMH NIH HHS/United States
- R01 NS049261/NS/NINDS NIH HHS/United States
- P01 HD035465/HD/NICHD NIH HHS/United States
- U19 HD035469-10/HD/NICHD NIH HHS/United States
- U01 HG004422-02/HG/NHGRI NIH HHS/United States
- MH066673/MH/NIMH NIH HHS/United States
- R37 MH057881/MH/NIMH NIH HHS/United States
- NS049261/NS/NINDS NIH HHS/United States
- P01 CA089392-08/CA/NCI NIH HHS/United States
- R01 MH081754-01/MH/NIMH NIH HHS/United States
- U19 HD035469-09/HD/NICHD NIH HHS/United States
- 075491/Z/04/WT_/Wellcome Trust/United Kingdom
- MH55284/MH/NIMH NIH HHS/United States
- R01 MH055284-04/MH/NIMH NIH HHS/United States
- MH080647/MH/NIMH NIH HHS/United States
- MC_U137761446/MRC_/Medical Research Council/United Kingdom
- R01 NS042165-05/NS/NINDS NIH HHS/United States
- MH061009/MH/NIMH NIH HHS/United States
- HD055782/HD/NICHD NIH HHS/United States
- MH081754/MH/NIMH NIH HHS/United States
- P50 HD055748-03/HD/NICHD NIH HHS/United States
- G0601030/MRC_/Medical Research Council/United Kingdom
- R01 MH061009/MH/NIMH NIH HHS/United States
- U19 HD035469/HD/NICHD NIH HHS/United States
- P50 HD055748-01/HD/NICHD NIH HHS/United States
- P01 CA089392/CA/NCI NIH HHS/United States
- T32 MH065215/MH/NIMH NIH HHS/United States
- CAPMC/ CIHR/Canada
- AS2077/AS/Autism Speaks/United States
- U10 MH066766/MH/NIMH NIH HHS/United States
- R01 NS042165/NS/NINDS NIH HHS/United States
- MH66766/MH/NIMH NIH HHS/United States
- MH52708/MH/NIMH NIH HHS/United States
- R01 MH057881/MH/NIMH NIH HHS/United States
- NS042165/NS/NINDS NIH HHS/United States
- P50 HD055748/HD/NICHD NIH HHS/United States
- AS7462/AS/Autism Speaks/United States
- U01 HG004422/HG/NHGRI NIH HHS/United States
- R01 DA019963-02/DA/NIDA NIH HHS/United States
- P01 NS026630-15/NS/NINDS NIH HHS/United States
- R01 DA013423-05/DA/NIDA NIH HHS/United States
- P50 HD055751/HD/NICHD NIH HHS/United States
- R01 MH052708-05/MH/NIMH NIH HHS/United States
- R01 DA019963/DA/NIDA NIH HHS/United States
- R01 DA013423/DA/NIDA NIH HHS/United States
- U54 MH066673-05/MH/NIMH NIH HHS/United States
- P50 HD055784/HD/NICHD NIH HHS/United States
- P50 HD055751-01/HD/NICHD NIH HHS/United States
- U10 MH066766-05/MH/NIMH NIH HHS/United States
- R01 MH080647/MH/NIMH NIH HHS/United States
- HD055784/HD/NICHD NIH HHS/United States
- P01 HD035465-01S1/HD/NICHD NIH HHS/United States
- R01 MH081754/MH/NIMH NIH HHS/United States
- R01 MH055284/MH/NIMH NIH HHS/United States
- R01 MH057881-02/MH/NIMH NIH HHS/United States
- UL1 TR000448/TR/NCATS NIH HHS/United States
- NS026630/NS/NINDS NIH HHS/United States
- HD35465/HD/NICHD NIH HHS/United States
- U54 MH066673/MH/NIMH NIH HHS/United States
- R01 NS049261-02/NS/NINDS NIH HHS/United States
- U19 HD035469-08/HD/NICHD NIH HHS/United States
- U19 HD035469-07/HD/NICHD NIH HHS/United States
- HD055751/HD/NICHD NIH HHS/United States
- P50 HD055782-04/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases