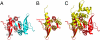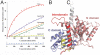Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition - PubMed (original) (raw)
Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition
Lisheng Ni et al. Proc Natl Acad Sci U S A. 2010.
Abstract
The segregation of plasmid DNA typically requires three elements: a DNA centromere site, an NTPase, and a centromere-binding protein. Because of their simplicity, plasmid partition systems represent tractable models to study the molecular basis of DNA segregation. Unlike eukaryotes, which utilize the GTPase tubulin to segregate DNA, the most common plasmid-encoded NTPases contain Walker-box and actin-like folds. Recently, a plasmid stability cassette on Bacillus thuringiensis pBtoxis encoding a putative FtsZ/tubulin-like NTPase called TubZ and DNA-binding protein called TubR has been described. How these proteins collaborate to impart plasmid stability, however, is unknown. Here we show that the TubR structure consists of an intertwined dimer with a winged helix-turn-helix (HTH) motif. Strikingly, however, the TubR recognition helices mediate dimerization, making canonical HTH-DNA interactions impossible. Mutagenesis data indicate that a basic patch, encompassing the two wing regions and the N termini of the recognition helices, mediates DNA binding, which indicates an unusual HTH-DNA interaction mode in which the N termini of the recognition helices insert into a single DNA groove and the wings into adjacent DNA grooves. The TubZ structure shows that it is as similar structurally to eukaryotic tubulin as it is to bacterial FtsZ. TubZ forms polymers with guanine nucleotide-binding characteristics and polymer dynamics similar to tubulin. Finally, we show that the exposed TubZ C-terminal region interacts with TubR-DNA, linking the TubR-bound pBtoxis to TubZ polymerization. The combined data suggest a mechanism for TubZ-polymer powered plasmid movement.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
B. thuringiensis pBtoxis TubR structure. (A) One TubR subunit is red and the other cyan. Secondary structural elements and N and C termini are labeled. (B) Superimposition of one subunit of TubR (Red) onto a S. aureus CzrA subunit (Yellow). Regions with different structures are labeled. (C) Same superimposition as B showing the location of the other subunit in the TubR and CzrA dimers after one subunit is overlaid. A_–_C are in the same orientation to highlight differences. Figs. 1 A_–_C, 2 C and D, 3 A, C, and E, 4_B_, and 5_B_ were made by using PyMOL (31).
Fig. 5.
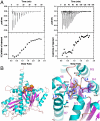
TubZ-guanine nucleotide interactions. (A) ITC binding isotherms showing TubZ-GDP (Left) and TubZ-GTP-_γ_-S interaction (Right). (B) Left: Overall structure of the TubZ-GTP-_γ_-S complex. β-strands are colored magenta and helices cyan, and the GTP-_γ_-S molecule is shown as cpk. Right: Close-up view of the GTP binding pocket with the initial _F_o-_F_c electron density map (Blue Mesh), contoured at 4.5σ, and calculated before the GTP-_γ_-S was included in refinement.
Fig. 4.
TubZ interacts with TubR-DNA and contains a tubulin/FtsZ fold. (A) FP assay measuring binding of FL TubZ, TubZ(1-470), TubZ(1-460), TubZ(1-442), and TubZ(1-407) to TubR-DNA. Below is the control (TubZ titrated into DNA alone). Millipolarization units and TubZ concentration (nM) are along the y and x axes, respectively. (B) TubZ(1-428) structure. The N domain or GTP-binding domain is colored salmon and the C domain purple. The interdomain helix, H7, is red. TubZ also contains an N-terminal helix, H0 (Yellow), and a C-terminal helix, H11 (White).
Fig. 3.
TubR-DNA binding. (A) Electrostatic surface potential of the TubR dimer. Blue and red represent electropositive and electronegative regions, respectively. (Left) The electronegative side of the TubR dimer, and the side on the right is the electropositive side. Labeled on the left side are the locations of the mutated residues. (B) FP binding isotherms showing the DNA binding of WT TubR and the K43A, R74A, R77A, and K79A mutants. Fluorescence polarization units (millipolarization) and TubR concentration (nM) are along the y and x axes, respectively. (C) TubR-DNA model showing TubR electrostatic potential. (D) Stoichiometry of TubR (subunit) binding showing titration curve of TubR into the 48-mer iteron resulting in a molar ratio of TubR subunit to DNA of eight (or four) dimers. (E) Left: Ribbon diagram of the TubR-DNA model with the recognition helices colored yellow. Right: Ribbon diagram showing a canonical HTH–DNA interaction (the λ repressor-DNA complex) with the recognition helices colored yellow (32).
Fig. 2.
The TubR “recognition helix” dimer. (A) Gel filtration studies on TubR mutants S63W and S63R showing that the S63W mutant remains dimeric whereas S63R is > 80% monomer. (B) Fluorescence polarization studies examining the ability of WT TubR, S63R, and S63W TubR mutants to bind iteronic DNA. Fluorescence polarization units (millipolarization) and TubR concentration (nM) are along the y and x axes, respectively. The _K_d of WT TubR for the centromere DNA is 8 ± 2 nm. (C) Superimposition of WT TubR (Green) onto the TubR S63W mutant structure (Tan). (D) Close-up of the site of the S63W mutation in the expanded TubR S63W dimer showing stacking interactions between the twofold related tryptophans.
Fig. 6.
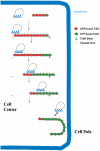
pBtoxis DNA partition model. In the first step, TubR, which is bound to its centromere on one of the replicated pBtoxis plasmids, contacts the TubZ C-terminal region (indicated by lines pointing from the TubZ “circles”) in a treadmilling TubZ polymer. TubZ subunits are lost from the - end and are added to the + end. TubR is pulled along the growing polymer by its TubR-TubZ interaction until it reaches the cell pole and is knocked off when it comes into contact with the membrane at the cell pole. TubZ reverses direction and may pick up the other TubR-pBtoxis complex and deliver it similarly to the opposite cell pole.
Comment in
- One-way ticket to the cell pole: plasmid transport by the prokaryotic tubulin homolog TubZ.
Barillà D. Barillà D. Proc Natl Acad Sci U S A. 2010 Jul 6;107(27):12061-2. doi: 10.1073/pnas.1007331107. Epub 2010 Jun 28. Proc Natl Acad Sci U S A. 2010. PMID: 20616084 Free PMC article. No abstract available.
Similar articles
- Filament formation of the FtsZ/tubulin-like protein TubZ from the Bacillus cereus pXO1 plasmid.
Hoshino S, Hayashi I. Hoshino S, et al. J Biol Chem. 2012 Sep 14;287(38):32103-12. doi: 10.1074/jbc.M112.373803. Epub 2012 Jul 30. J Biol Chem. 2012. PMID: 22847006 Free PMC article. - Cooperative DNA Binding of the Plasmid Partitioning Protein TubR from the Bacillus cereus pXO1 Plasmid.
Hayashi I, Oda T, Sato M, Fuchigami S. Hayashi I, et al. J Mol Biol. 2018 Dec 7;430(24):5015-5028. doi: 10.1016/j.jmb.2018.11.001. Epub 2018 Nov 8. J Mol Biol. 2018. PMID: 30414406 - Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis.
Larsen RA, Cusumano C, Fujioka A, Lim-Fong G, Patterson P, Pogliano J. Larsen RA, et al. Genes Dev. 2007 Jun 1;21(11):1340-52. doi: 10.1101/gad.1546107. Epub 2007 May 17. Genes Dev. 2007. PMID: 17510284 Free PMC article. - Structural biology of plasmid partition: uncovering the molecular mechanisms of DNA segregation.
Schumacher MA. Schumacher MA. Biochem J. 2008 May 15;412(1):1-18. doi: 10.1042/BJ20080359. Biochem J. 2008. PMID: 18426389 Review. - Tubulin-Like Proteins in Prokaryotic DNA Positioning.
Fink G, Aylett CHS. Fink G, et al. Subcell Biochem. 2017;84:323-356. doi: 10.1007/978-3-319-53047-5_11. Subcell Biochem. 2017. PMID: 28500531 Review.
Cited by
- β-Strand-mediated interactions of protein domains.
Bhat AS, Kinch LN, Grishin NV. Bhat AS, et al. Proteins. 2020 Nov;88(11):1513-1527. doi: 10.1002/prot.25970. Epub 2020 Jul 11. Proteins. 2020. PMID: 32543729 Free PMC article. - Segrosome Complex Formation during DNA Trafficking in Bacterial Cell Division.
Oliva MA. Oliva MA. Front Mol Biosci. 2016 Sep 9;3:51. doi: 10.3389/fmolb.2016.00051. eCollection 2016. Front Mol Biosci. 2016. PMID: 27668216 Free PMC article. Review. - Filament structure of bacterial tubulin homologue TubZ.
Aylett CH, Wang Q, Michie KA, Amos LA, Löwe J. Aylett CH, et al. Proc Natl Acad Sci U S A. 2010 Nov 16;107(46):19766-71. doi: 10.1073/pnas.1010176107. Epub 2010 Oct 25. Proc Natl Acad Sci U S A. 2010. PMID: 20974911 Free PMC article. - One-way ticket to the cell pole: plasmid transport by the prokaryotic tubulin homolog TubZ.
Barillà D. Barillà D. Proc Natl Acad Sci U S A. 2010 Jul 6;107(27):12061-2. doi: 10.1073/pnas.1007331107. Epub 2010 Jun 28. Proc Natl Acad Sci U S A. 2010. PMID: 20616084 Free PMC article. No abstract available. - The structure and assembly mechanism of a novel three-stranded tubulin filament that centers phage DNA.
Zehr EA, Kraemer JA, Erb ML, Coker JK, Montabana EA, Pogliano J, Agard DA. Zehr EA, et al. Structure. 2014 Apr 8;22(4):539-48. doi: 10.1016/j.str.2014.02.006. Epub 2014 Mar 13. Structure. 2014. PMID: 24631461 Free PMC article.
References
- Nogales E, Wolf SG, Downing KH. Structure of the αβ tubulin dimer by electron crystallography. Nature. 1998;391:199–203. - PubMed
- Nogales E, Whittaker M, Milligan RA, Downing KH. High-resolution model of the microtubule. Cell. 1999;96:79–88. - PubMed
- Nogales E. Structural insights into microtubule function. Annu Rev Biochem. 2000;69:277–302. - PubMed
- Oliva MA, Cordell SC, Löwe J. Structural insights into FtsZ protofilament formation. Nat Struct Mol Biol. 2004;11:1243–1250. - PubMed
- Oliva MA, Trambaiolo D, Löwe J. Structural insights into the conformational variability of FtsZ. J Mol Biol. 2007;373:1229–1242. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases