A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids - PubMed (original) (raw)
A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids
Jan Janouskovec et al. Proc Natl Acad Sci U S A. 2010.
Abstract
The discovery of a nonphotosynthetic plastid in malaria and other apicomplexan parasites has sparked a contentious debate about its evolutionary origin. Molecular data have led to conflicting conclusions supporting either its green algal origin or red algal origin, perhaps in common with the plastid of related dinoflagellates. This distinction is critical to our understanding of apicomplexan evolution and the evolutionary history of endosymbiosis and photosynthesis; however, the two plastids are nearly impossible to compare due to their nonoverlapping information content. Here we describe the complete plastid genome sequences and plastid-associated data from two independent photosynthetic lineages represented by Chromera velia and an undescribed alga CCMP3155 that we show are closely related to apicomplexans. These plastids contain a suite of features retained in either apicomplexan (four plastid membranes, the ribosomal superoperon, conserved gene order) or dinoflagellate plastids (form II Rubisco acquired by horizontal transfer, transcript polyuridylylation, thylakoids stacked in triplets) and encode a full collective complement of their reduced gene sets. Together with whole plastid genome phylogenies, these characteristics provide multiple lines of evidence that the extant plastids of apicomplexans and dinoflagellates were inherited by linear descent from a common red algal endosymbiont. Our phylogenetic analyses also support their close relationship to plastids of heterokont algae, indicating they all derive from the same endosymbiosis. Altogether, these findings support a relatively simple path of linear descent for the evolution of photosynthesis in a large proportion of algae and emphasize plastid loss in several lineages (e.g., ciliates, Cryptosporidium, and Phytophthora).
Conflict of interest statement
The authors declare no conflict of interest.
Figures
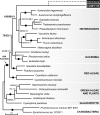
Fig. 1.
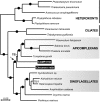
The nuclear phylogeny of C. velia and CCMP3155 show they are two independent photosynthetic lineages closely related to apicomplexan parasites (arrow). The RAxML tree is derived from concatenation of eight nuclear encoded genes (7,137 characters). RAxML/ MrBayes supports are shown above branches; solid circles indicate 100/1 supports.
Fig. 2.
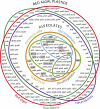
Venn diagram of plastid genome contents in various red plastid lineages. Overlap between the four lineages of alveolates represented by the center rings reveals that plastids of C. velia and CCMP3155 collectively encode all genes found in both apicoplasts and dinoflagellate plastids. Gray boxes highlight 18 genes that are absent in plastids of plants and green algae but all found in alveolate and red algal derived plastids. Genes that originated through horizontal gene transfer are marked with an asterisk. The diagram does not include genes for tRNAs, other small RNAs (5S rRNA, ffs, tmRNAs, rnpB) and the ppd gene horizontally transferred to the CCMP3155 plastid genome.
Fig. 3.
Form II Rubisco and polyU tails in plastid mRNA link the plastids of C. velia, CCMP3155, and dinoflagellates. (A) RaxML phylogenetic tree of form II Rubisco shows the C. velia and CCMP3155 form II Rubisco genes are closely related to homologs in dinoflagellates. (B) 3′ tails of C. velia plastid mRNAs, represented here by the psaA transcript, contain short polyU tails absent from plastid DNA, a feature previously considered unique to dinoflagellate plastids. Transcripts from other genes are shown in
Fig. S6
. Underlined thymidines may correspond to the 5′UTRs of the circularized transcripts (Materials and Methods).
Fig. 4.
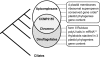
Summary of plastid evolution in alveolates. The plastid genomes of C. velia and CCMP3155 provide a direct link between the plastids of apicomplexans and dinoflagellates because they retain ancestral features that were previously thought to be exclusive to one or the other of these lineages (boxed at the right). Relationships between the lineages based on nuclear data are shown at the left. An asterisk indicates that several regions of conserved gene order are found between the plastid genomes of apicomplexans and CCMP3155, and CCMP3155 and C. velia. Double asterisk indicates that the presence of polyUs in CCMP3155 plastid transcripts has not yet been determined.
Fig. 5.
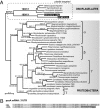
Plastid phylogenies of 34 conserved plastid genes confirm that CCMP3155 contains a red algal plastid and strongly support its close relationship to plastids of heterokonts. The dotted branch indicates the placement of C. velia sequence, which received complete support in all analyses, but was excluded because of its high rate of substitution. The RAxML tree shows RAxML/PhyML-CAT/MrBayes/PhyloBayes supports over branches; solid circles indicate 100/100/1/1 support. Only ≥60/≥50/≥0.98/≥0.98 branch supports are shown as significant.
Similar articles
- Chromera velia, endosymbioses and the rhodoplex hypothesis--plastid evolution in cryptophytes, alveolates, stramenopiles, and haptophytes (CASH lineages).
Petersen J, Ludewig AK, Michael V, Bunk B, Jarek M, Baurain D, Brinkmann H. Petersen J, et al. Genome Biol Evol. 2014 Mar;6(3):666-84. doi: 10.1093/gbe/evu043. Genome Biol Evol. 2014. PMID: 24572015 Free PMC article. - Red and problematic green phylogenetic signals among thousands of nuclear genes from the photosynthetic and apicomplexa-related Chromera velia.
Woehle C, Dagan T, Martin WF, Gould SB. Woehle C, et al. Genome Biol Evol. 2011;3:1220-30. doi: 10.1093/gbe/evr100. Epub 2011 Sep 28. Genome Biol Evol. 2011. PMID: 21965651 Free PMC article. - Nuclear-encoded, plastid-targeted genes suggest a single common origin for apicomplexan and dinoflagellate plastids.
Fast NM, Kissinger JC, Roos DS, Keeling PJ. Fast NM, et al. Mol Biol Evol. 2001 Mar;18(3):418-26. doi: 10.1093/oxfordjournals.molbev.a003818. Mol Biol Evol. 2001. PMID: 11230543 - Evolution of the apicoplast and its hosts: from heterotrophy to autotrophy and back again.
Oborník M, Janouskovec J, Chrudimský T, Lukes J. Oborník M, et al. Int J Parasitol. 2009 Jan;39(1):1-12. doi: 10.1016/j.ijpara.2008.07.010. Epub 2008 Sep 12. Int J Parasitol. 2009. PMID: 18822291 Review. - The endosymbiotic origin, diversification and fate of plastids.
Keeling PJ. Keeling PJ. Philos Trans R Soc Lond B Biol Sci. 2010 Mar 12;365(1541):729-48. doi: 10.1098/rstb.2009.0103. Philos Trans R Soc Lond B Biol Sci. 2010. PMID: 20124341 Free PMC article. Review.
Cited by
- Eye-like ocelloids are built from different endosymbiotically acquired components.
Gavelis GS, Hayakawa S, White RA 3rd, Gojobori T, Suttle CA, Keeling PJ, Leander BS. Gavelis GS, et al. Nature. 2015 Jul 9;523(7559):204-7. doi: 10.1038/nature14593. Epub 2015 Jul 1. Nature. 2015. PMID: 26131935 - Volatiles of the Apicomplexan Alga Chromera velia and Associated Bacteria.
Koteska D, Marter P, Huang S, Pradella S, Petersen J, Schulz S. Koteska D, et al. Chembiochem. 2023 Feb 1;24(3):e202200530. doi: 10.1002/cbic.202200530. Epub 2022 Dec 20. Chembiochem. 2023. PMID: 36416092 Free PMC article. - After the primary endosymbiosis: an update on the chromalveolate hypothesis and the origins of algae with Chl c.
Green BR. Green BR. Photosynth Res. 2011 Jan;107(1):103-15. doi: 10.1007/s11120-010-9584-2. Epub 2010 Jul 30. Photosynth Res. 2011. PMID: 20676772 Review. - The search for the missing link: a relic plastid in Perkinsus?
Fernández Robledo JA, Caler E, Matsuzaki M, Keeling PJ, Shanmugam D, Roos DS, Vasta GR. Fernández Robledo JA, et al. Int J Parasitol. 2011 Oct;41(12):1217-29. doi: 10.1016/j.ijpara.2011.07.008. Epub 2011 Aug 22. Int J Parasitol. 2011. PMID: 21889509 Free PMC article. Review. - Evolution of chloroplast transcript processing in Plasmodium and its chromerid algal relatives.
Dorrell RG, Drew J, Nisbet RE, Howe CJ. Dorrell RG, et al. PLoS Genet. 2014 Jan;10(1):e1004008. doi: 10.1371/journal.pgen.1004008. Epub 2014 Jan 16. PLoS Genet. 2014. PMID: 24453981 Free PMC article.
References
- McFadden GI, Reith ME, Munholland J, Lang-Unnasch N. Plastid in human parasites. Nature. 1996;381:482. - PubMed
- Wilson RJ, et al. Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J Mol Biol. 1996;261:155–172. - PubMed
- Zhang Z, Green BR, Cavalier-Smith T. Single gene circles in dinoflagellate chloroplast genomes. Nature. 1999;400:155–159. - PubMed
- Köhler S, et al. A plastid of probable green algal origin in Apicomplexan parasites. Science. 1997;275:1485–1489. - PubMed
- Cai X, Fuller AL, McDougald LR, Zhu G. Apicoplast genome of the coccidian Eimeria tenella. Gene. 2003;321:39–46. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials