Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis - PubMed (original) (raw)
Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis
Thomas A Milne et al. Mol Cell. 2010.
Abstract
MLL1 fusion proteins activate HoxA9 gene expression and cause aggressive leukemias that respond poorly to treatment, but how they recognize and stably bind to HoxA9 is not clearly understood. In a systematic analysis of MLL1 domain recruitment activity, we identified an essential MLL1 recruitment domain that includes the CXXC domain and PHD fingers and is controlled by direct interactions with the PAF elongation complex and H3K4Me2/3. MLL1 fusion proteins lack the PHD fingers and require prebinding of a wild-type MLL1 complex and CXXC domain recognition of DNA for stable HoxA9 association. Together, these results suggest that specific recruitment of MLL1 requires multiple interactions and is a precondition for stable recruitment of MLL1 fusion proteins to HoxA9 in leukemogenesis. Since wild-type MLL1 and oncogenic MLL1 fusion proteins have overlapping yet distinct recruitment mechanisms, this creates a window of opportunity that could be exploited for the development of targeted therapies.
Copyright (c) 2010 Elsevier Inc. All rights reserved.
Figures
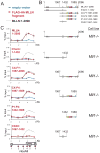
Figure 1. Recruitment of the MLL1 complex is required for HoxA9 activation
(A) Important MLL1 domains and protein interactions. The MLL1 protein is proteolytically cleaved in the cell into N terminal (MLL1N) and C terminal (MLL1C) proteins. All MLL1 leukemogenic fusions break just before the first PHD finger (line, Leukemia Breakpoint). Two MLL1 fusion proteins MLL-AF9 and MLL-ENL are shown as examples. (B) HoxA9 protein expression depends on the expression of MLL1. Extracts were prepared from Mll1_−/_− mouse embryonic fibroblast (MEF) knockout cells (lane −) and Mll1_−/_− MEF cells that have been reconstituted with full length MLL1 (lane +). Western blots were probed with the antibodies indicated. (C) Chromatin immunoprecipitation (ChIP) experiments at HoxA9 in Mll1_−/_− (blue line) versus Mll1_−/_− reconstituted with MLL1 (red line) MEF cells using the antibodies indicated. A schematic of the mouse HoxA9 locus with red boxes indicating the positions of 6 primer/probe sets is shown at the bottom of the ChIP figures. ChIP results shown are typical for at least two independent experiments. Error bars represent standard deviation of three PCR reactions. See also Figure S1.
Figure 2. The CXXC domain and PHD fingers are sufficient for targeting MLL1 to the HoxA9 locus in vivo
(A) Legend for the ChIP experiments in C. Blue line= ChIP in cells transfected with an empty vector, Red line= ChIP in cells transfected with the constructs indicated and Grey dots = the binding pattern of full length MLLN for comparison. (B) Schematics showing the series of deletion constructs. (C) The regions containing the MLL1 CXXC domain and the PHD fingers are both necessary for targeting MLL1 to the HoxA9 locus. Different fragments of the MLL1 N terminus were FLAG and HA (fh) double tagged and expressed in Mll1_−/_− MEFs and α-HA ChIP experiments were performed. ChIP results shown are typical for at least three independent experiments. Error bars represent standard deviation of three separate PCR reactions. See also Figure S2.
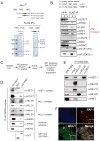
Figure 3. The MLL1 complex interacts specifically with the PAF elongation complex
(A) 293 cells stably expressing either FLAG-HA tagged (fh) CXXC:1067–1432 or fh-P1-P4:1432–1989 were subjected to a one step FLAG purification. Mass spectroscopy analysis identified components of the MLL1 core complex (HCFC1 and HCFC2) and components of PAF1C (shown in red) (B) FLAG IP’s were done from 293 cell nuclear extracts expressing either an empty vector (1), fh-CXXC:1067–1432 (2) or fh- P1-P4:1432–1989 (3). Western blots were probed with the antibodies indicated. (C) Scheme for purification of multiple MLL family complexes from HeLa extracts expressing a FLAG (f-) tagged WDR5 protein. (D) GST-PAF1C specifically interacts with components of the MLL1 complex. The f-WDR5 preparation was subjected to GST pulldowns in the presence of either GST or a GST-hPAF1C and the results were blotted and probed with the antibodies indicated. (E) PAF1 interacts with the MLL1 complex in vivo. 293 nuclear extracts were used for IP’s with the antibodies indicated and western blots were probed with the antibodies indicated. (F) Immunofluorescence of PAF1 (red) and MLL1 (green) in 293 cells. PAF1 and MLL1 overlap in only discrete regions, a blowup of a region of overlap is shown as an example (white box). The white bar represents 5μm. See also Figure S3.
Figure 4. The MLL1 CXXC/RD1 domain interacts directly with PAF1 and PHD finger 3 interacts directly with H3K4Me2/3
(A) PAF1C interacts directly with the CXXC containing RD1 fragment of MLL1. The MLL1 CXXC region was divided into two fragments (RD1 and RD2) and expressed as recombinant GST proteins. Purified GST (1), GST-RD1 (2) and GST-RD2 (3) were used in GST pulldown experiments with a recombinant purified PAF1C. Westerns were probed with the antibodies indicated. (B) The PAF1 component of PAF1C interacts directly with MLL RD1. GST-RD1 was used in a GST pulldown experiment with individual FLAG (f-) tagged components of human PAF1C as shown. The upper panel is a gel stained with colloidal blue and the lower panels are western blots of the same pulldowns using the antibodies indicated. (C) Only full length MLL1-RD1 interacts efficiently with PAF1, while MLL2-RD1 does not. A series of deletions of the MLL1 RD1 region were made and subjected to GST pulldowns with purified f-PAF1. Westerns were probed with the antibodies indicated. (D) Top: conservation with MLL2 and the positions of point mutations made in the MLL1 CXXC domain are shown. Bottom: The R1153A point mutation disrupts the direct interaction between purified GST-RD1 and purified f-PAF1 protein. GST pulldowns were done as in (A). (E) PHD fingers 1 to 4 with and without a point mutation in PHD3 (W1594E) were FLAG and HA double tagged and expressed in 293 cells. Nuclear extracts were subjected to peptide pulldowns as indicated and probed with the antibodies shown. (F) Recombinantly expressed and purified dual PHD3/Bromodomain of MLL1 binds specifically to H3 lysine 4 di and trimethyl peptides (top panel). A W1594E point mutation in PHD3 abolishes this interaction (bottom panel). See also Figure S4.
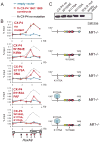
Figure 5. Binding to H3K4Me and PAF1 are both necessary for recruitment of MLL1 across HoxA9 in vivo
(A) Legend for the ChIP experiments in C. Blue line= HA ChIP in cells transfected with an empty vector, Red line= HA ChIP in cells transfected with the CX-P4 constructs indicated and Grey dots = the binding pattern of CX-P4 with no mutation. (B) PAF1 and H3K4Me interactions are important for recruiting MLL1 to HoxA9. Using α-HA ChIP, recruitment of the MLL1 fh-CX-P4 protein to HoxA9 in Mll1_−/_− MEF cells (no mutant) was compared to individual PHD3 W1594E, K1176A and R1153A point mutations and a triple mutant. ChIP results shown are typical for at least two independent experiments. Error bars represent standard deviation of three separate PCR reactions. (C) An α-HA western blot showing expression of the wild type and mutant constructs from the experiments in B. W,K,R mut = the W1594E, K1176A, R1153A triple mutant. See also Figure S5.
Figure 6. The MLL-AF9 fusion protein needs wild type MLL1, PAF1 and DNA binding for recruitment to HoxA9
(A) Legend for the ChIP experiments in B-J. Blue line= HA ChIP in cells transfected with an empty vector, Red line= HA ChIP in cells transfected with the constructs indicated and Grey dots = the binding pattern of fh-MLL-AF9 in Mll1_−/_− + MLL cells for comparison. (B) – (J) α-HA ChIP across HoxA9 for the constructs and in the specific cell lines as indicated. ChIP results shown are typical for at least three independent experiments. Error bars represent standard deviation of three separate PCR reactions. See also Figure S6.
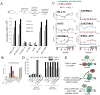
Figure 7. Recruitment of fh-MLL-AF9 to HoxA9 in bone marrow cells is essential for leukemic transformation
(A) Leukemogenic transformation of MLL-AF9 constructs. Retrovirally expressed FLAG and HA tagged (fh-) MLL-AF9 fusion protein constructs that were either normal for MLL-AF9 or had R1153A (PAF1 disruption), or K1176A (DNA binding disruption) point mutations were transduced into murine bone marrow cells, along with empty vector, untagged MLL-ENL, and triple FLAG tagged NUP98-Jarid controls. Error bars represent the standard deviation of averaged, cumulative cell numbers across five independent lines for each construct. (B) HoxA9 expression in transformed and control cell lines. Cells from (A) were tested for HoxA9 expression and the average values across all five independent cell lines per construct are shown. Error bars represent standard deviation. HoxA9 expression in Mll1_−/_− and Mll1_−/_− + MLL cell lines are also shown for comparison. (C) ChIP in progenitor enriched (Blue line) and fh-MLLAF9 (line #1, Red line) cells with the antibodies indicated. ChIP experiments in Mll1_−/_− + MLL cells are shown for comparison (Grey dots). (D) PAF1 is required for MLL1 and MLL-AF9 binding to HoxA9. The fh-MLLAF9#1 cell line was treated with control vs PAF1 siRNA’s and the cells were subjected to western blot (inset) and ChIP analysis using the antibodies indicated. α-HA antibody recognizes the HA tagged MLL-AF9 fusion protein. ChIP results were quantified using a primer/probe set for the silent Hoxc8 locus or the mouse HoxA9 coding region (primer/probe set 5 from D). ChIP signal was quantified to inputs and then relative to the HoxA9 control signal which was arbitrarily set to 100. (E) 1= wild type MLL1 is recruited to HoxA9 through interactions with H3K4Me, PAF1 and DNA. 2= MLL1-mediated activation of the HoxA9 locus disrupts binding of repressors such as ESET and creates a more “open” chromatin conformation. 3= An MLL1 fusion protein can now bind through interactions with PAF1 and with CpG rich DNA. ChIP results shown are typical for at least two independent experiments. Error bars represent standard deviation of three separate PCR reactions. See also Figure S7.
Comment in
- Leukaemia: MLL makes friends and influences.
McCarthy N. McCarthy N. Nat Rev Cancer. 2010 Aug;10(8):529. doi: 10.1038/nrc2904. Nat Rev Cancer. 2010. PMID: 20677349 No abstract available.
Similar articles
- NUP98 Fusion Proteins Interact with the NSL and MLL1 Complexes to Drive Leukemogenesis.
Xu H, Valerio DG, Eisold ME, Sinha A, Koche RP, Hu W, Chen CW, Chu SH, Brien GL, Park CY, Hsieh JJ, Ernst P, Armstrong SA. Xu H, et al. Cancer Cell. 2016 Dec 12;30(6):863-878. doi: 10.1016/j.ccell.2016.10.019. Epub 2016 Nov 23. Cancer Cell. 2016. PMID: 27889185 Free PMC article. - Functional specificity of CpG DNA-binding CXXC domains in mixed lineage leukemia.
Risner LE, Kuntimaddi A, Lokken AA, Achille NJ, Birch NW, Schoenfelt K, Bushweller JH, Zeleznik-Le NJ. Risner LE, et al. J Biol Chem. 2013 Oct 11;288(41):29901-10. doi: 10.1074/jbc.M113.474858. Epub 2013 Aug 29. J Biol Chem. 2013. PMID: 23990460 Free PMC article. - The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis.
Muntean AG, Tan J, Sitwala K, Huang Y, Bronstein J, Connelly JA, Basrur V, Elenitoba-Johnson KS, Hess JL. Muntean AG, et al. Cancer Cell. 2010 Jun 15;17(6):609-21. doi: 10.1016/j.ccr.2010.04.012. Cancer Cell. 2010. PMID: 20541477 Free PMC article. - Deregulation of the HOXA9/MEIS1 axis in acute leukemia.
Collins CT, Hess JL. Collins CT, et al. Curr Opin Hematol. 2016 Jul;23(4):354-61. doi: 10.1097/MOH.0000000000000245. Curr Opin Hematol. 2016. PMID: 27258906 Free PMC article. Review. - MLL1/WDR5 complex in leukemogenesis and epigenetic regulation.
Wu M, Shu HB. Wu M, et al. Chin J Cancer. 2011 Apr;30(4):240-6. doi: 10.5732/cjc.011.10055. Chin J Cancer. 2011. PMID: 21439245 Free PMC article. Review.
Cited by
- Histone H2B ubiquitin ligase RNF20 is required for MLL-rearranged leukemia.
Wang E, Kawaoka S, Yu M, Shi J, Ni T, Yang W, Zhu J, Roeder RG, Vakoc CR. Wang E, et al. Proc Natl Acad Sci U S A. 2013 Mar 5;110(10):3901-6. doi: 10.1073/pnas.1301045110. Epub 2013 Feb 14. Proc Natl Acad Sci U S A. 2013. PMID: 23412334 Free PMC article. - SET1/MLL family of proteins: functions beyond histone methylation.
Sugeedha J, Gautam J, Tyagi S. Sugeedha J, et al. Epigenetics. 2021 May;16(5):469-487. doi: 10.1080/15592294.2020.1809873. Epub 2020 Aug 31. Epigenetics. 2021. PMID: 32795105 Free PMC article. Review. - Menin as a hub controlling mixed lineage leukemia.
Thiel AT, Huang J, Lei M, Hua X. Thiel AT, et al. Bioessays. 2012 Sep;34(9):771-80. doi: 10.1002/bies.201200007. Epub 2012 Jul 24. Bioessays. 2012. PMID: 22829075 Free PMC article. Review. - Leukaemia: MLL makes friends and influences.
McCarthy N. McCarthy N. Nat Rev Cancer. 2010 Aug;10(8):529. doi: 10.1038/nrc2904. Nat Rev Cancer. 2010. PMID: 20677349 No abstract available. - Effective Menin inhibitor-based combinations against AML with MLL rearrangement or NPM1 mutation (NPM1c).
Fiskus W, Boettcher S, Daver N, Mill CP, Sasaki K, Birdwell CE, Davis JA, Takahashi K, Kadia TM, DiNardo CD, Jin Q, Qi Y, Su X, McGeehan GM, Khoury JD, Ebert BL, Bhalla KN. Fiskus W, et al. Blood Cancer J. 2022 Jan 11;12(1):5. doi: 10.1038/s41408-021-00603-3. Blood Cancer J. 2022. PMID: 35017466 Free PMC article.
References
- Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001;20:5695–5707. - PubMed
- Bach C, Mueller D, Buhl S, Garcia-Cuellar MP, Slany RK. Alterations of the CXXC domain preclude oncogenic activation of mixed-lineage leukemia 2. Oncogene. 2009;28:815–823. - PubMed
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical