Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a - PubMed (original) (raw)
Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a
Kyoko L Yap et al. Mol Cell. 2010.
Abstract
Expression of the INK4b/ARF/INK4a tumor suppressor locus in normal and cancerous cell growth is controlled by methylation of histone H3 at lysine 27 (H3K27me) as directed by the Polycomb group proteins. The antisense noncoding RNA ANRIL of the INK4b/ARF/INK4a locus is also important for expression of the protein-coding genes in cis, but its mechanism has remained elusive. Here we report that chromobox 7 (CBX7) within the polycomb repressive complex 1 binds to ANRIL, and both CBX7 and ANRIL are found at elevated levels in prostate cancer tissues. In concert with H3K27me recognition, binding to RNA contributes to CBX7 function, and disruption of either interaction impacts the ability of CBX7 to repress the INK4b/ARF/INK4a locus and control senescence. Structure-guided analysis reveals the molecular interplay between noncoding RNA and H3K27me as mediated by the conserved chromodomain. Our study suggests a mechanism by which noncoding RNA participates directly in epigenetic transcriptional repression.
Copyright (c) 2010 Elsevier Inc. All rights reserved.
Figures
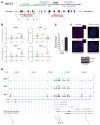
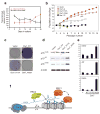
Figure 1. Non-coding RNA ANRIL is Up-regulated in Prostate Cancer Tissues and Coincides with CBX7 and EZH2 Expression
a, Schematic representation of the genomic organization of the INK4a/ARF gene cluster. Three genes in the INK4a/ARF locus are color-coded. Transcripts antisense to ANRIL (blue) used to inhibit ANRIL interactions (see Figure 2) are indicated above ANRIL. Roman numerals denote sites of quantitative ChIP as shown in Figure 3. b, Normalized expression levels of ANRIL, p16, CBX7 and EZH2 in normal prostate epithelium cells, and preneoplastic PIN and PCa prostate cancer cells. Each plotted point represents a separate tissue sample. c, Levels of ANRIL transcript relative to HPRT as detected in normal prostate epithelium and PC3 prostate cancer cells. d_,_ RNA-FISH (fluorescence in situ hybridization) of ANRIL as detected in normal prostate and prostate cancer (PCa) cells. The housekeeping genes GAPDH and HPRT are shown as input controls. e, Relative enrichment of RNA polymerase II, EZH2 and CBX7 associated with loci spanning the INK4a/ARF region in the IMR90 fibroblasts (red) or PC3 prostate carcinoma (green) background. Standard ChIP analysis was performed at selected points overlapping transcript starts identified within the UCSC Genome Browser (chr9: 21,710,518-2,301,141), based on RefSeq, Uniprot, GeneBank, CCDS, and Comparative Genomics and plotted using Genome Graphs (
). ChIP primers used are listed in Table S2. See also Figure S1.
Figure 2. Competitive inhibition of ANRIL transcript interactions results in loss of INK4a repression and limitation of cellular lifespan
a, Transcript antisense to the ANRIL long non-coding transcript relative to the INK/ARF locus (as indicated by arrows above ANRIL in Figure 1a) influence lifespan of normal IMR-90 diploid fibroblasts, transduced with retrovirus expressing each of the antisense cDNA as indicated, and quantified through population doublings following each passage (n=3). Points equal mean; bars equal standard deviation. b, Levels of p16INK4a, p15INK4b and tubulin were evaluated from retrovirally-transduced IMR-90 cell extracts following passage 20 by immunoblots. c, RNA transcript levels of INK4a and INK4b following passages as indicated. d, Deposition and occupation of histone methylation marks, H3K27me3 and H3K4me3, CBX7, and EZH2 at the transcription start site (position III) of the INK4a locus as measured by ChIP analysis. e, Competition study using antisense transcripts to ANRIL were generated by in vitro transcription and analyzed from nuclear proteins bound by RNA-EMSA.
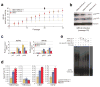
Figure 3. ANRIL ncRNA Associates Specifically with Members of the PRC1 Complex
a, Recovery of ANRIL transcripts by CLIP analysis of CBX7 in chromatin from asynchronously growing PC3 cells. Lane 1, genomic DNA prior to DNA hydrolysis from nuclei recovered; reverse transcription PCR (rtPCR) of heteronuclear RNA (lane 2) and cytoplasmic RNA (lane 3) recovered from PC3 cell nuclei; lane 4, DNA molecular ladder marker; RNA-ChIP of CBX7-bound RNA and rtPCR using antisense primer to detect sense strand of ANRIL transcript (lanes 5, 6) or sense primer to detect anti-sense strand (lanes 7, 8) in the absence or presence of AMV reverse transcriptase, respectively. b and c, FLAG-tagged CBX7 purified from human PC3 cells binds to the ANRIL transcript. Silver staining was used to visualize the human CBX7 complex purified from the PC3 cells. Identification of CBX7/PRC1 components was performed by MALDI-TOF mass spectrometry analysis and specific protein components confirmed (in c) by immunoblotting with antisera directed against RING6A/MBLR, CBX7, and HP1α. d, Band shift of the sense and anti-sense ANRIL transcript by the homogenous CBX7/PRC1 complex by RNA EMSA assay. 50 and 200 ng of purified PRC1 proteins were used to visualize the RNA binding potential of the CBX7-associated complex for the segment of the ncRNA transcript generated by in vitro transcription (lanes 2, 3). Competitive (50x and 1000x excess) unlabeled ncRNA transcript was used as shown to compete for ANRIL (lanes 4, 5). Note that anti-sense ANRIL does not form a stable complex with the purified PRC1 (lane 6). e, Total nuclear extract from PC3 cells was used to evaluate by Western blot analysis the association of PRC1 proteins CBX7, Bmi1 and HP1α with the biotinylated ANRIL transcript generated in d. See also Figure S2.
Figure 4. Recruitment of Polycomb Proteins to the INK4a/ARF Locus is Mediated by RNA Polymerase II Transcripts
a, Quantitative ChIP (qChIP) analysis from whole nuclei recovered from 3×109 PC3 cells following RNase A treatment, with or without RNase inhibitor added, at the sites noted in Figure 1a in Roman numerals (I to V) indicates the presence of CBX7, Bmi1, H3K27me3, PHC2 (polyhomeotic-like 2), Suz12 (Suppressor of Zeste 12 protein homolog), H2AK119Ub and histone H1. Controls are of mock treatment and levels of H3 are shown below. b, CBX7 occupation relative to histone H3 were measured with ChIP at the different sites along the INK4a/ARF locus and time points after PC3 cells treated with α-amanitin (10 μg/ml). c, In vitro transcription of ANRIL hnRNA transcripts from PC3 whole nuclei shows sensitivity to α-amanitin (upper panel). Exposure times for the transcripts are shown. Quantitative measurements of [α-32P]-UTP incorporation into nascent transcripts of ANRIL, HPRT and 5S RNA are plotted based on densitometry and liquid scintillation measurements of the [32P]-labeled transcripts (lower panel).
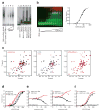
Figure 5. Molecular Interplay between _ANRIL_-derived RNA and H3K27me as mediated by the CBX7 chromodomain
a, EMSA showing CBX7 full-length (CBX7FL, left) and chromodomain (CBX7CD, right) binding to RNA. Increasing concentrations of CBX7FL are shown in the presence of a 26-nt RNA; CBX7CD and other chromodomains are shown in the presence of a 13-nt ssDNA, 13-nt RNA (RNA13) or 26-nt RNA (RNA26). b, Fluorescence-based EMSA of titration of CBX7CD (red) into a constant concentration of RNA (green) shows an increasing amount of CBX7CD-RNA complex formation (yellow). The binding curve (KD = ~51 μM) based on complex overlay signal is shown at right. c, [1H,15N]-HSQC spectra of CBX7CD in the presence of H3K27me2 peptide, 14-nt ssDNA, 14-nt loop RNA and RNA after treatment with RNase A. The resonances of several residues not observed in the HSQC underwent severe line broadening and were confirmed by 3D experiments. d, Binding curves for CBX7CD binding to fluorescein-labeled ANRIL loopC RNA (black), H3K27me3 (red) or H3K27me2 (blue) peptide, as monitored by fluorescence anisotropy. e, H3K27me3/RNA competition experiments, in which unlabeled competing ligand is titrated against constant concentrations of CBX7CD and fluorescein-labeled ligand. f, Binding curves for CBX7CD binding to fluorescein-labeled ANRIL loop C in the presence (black) or absence (red) of saturating amounts of H3K27me3 peptide. The KD values for these curves are 75.9 ± 11.2 μM and 56.3 ± 6.8 μM, respectively, yielding a negative cooperativity factor α of 1.3. All anisotropy data were measured in triplicate. See also Figures S1, S3 and S4.
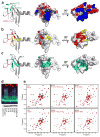
Figure 6. CBX7 Chromodomain Employs Distinct Regions and Residues for Binding H3K27me or RNA
a, Representative 3D NMR structure of CBX7CD in complex with H3K27me2 peptide depicted as ribbon or surface colored by Poisson-Boltzmann electrostatic potential, with aromatic cage residues as shown. b, CBX7CD structure with residues as determined by NMR titration depicting severe line broadening (red) or moderate peak shifts (yellow) upon binding H3K27me2 peptide. c, Residues undergoing perturbation upon RNA addition at early (CBX7CD:RNA molar ratio 1:0.2, blue) and later (1:0.8, cyan) points during titration. d, Fluorescence-based EMSA depicting 26-nt RNA binding to different point mutants of CBX7CD based on the NMR titration in b and c. e, [1H,15N]-HSQC spectra of wild-type and corresponding point mutants showing affected H3K27me2 or RNA binding. See also Figures S5 and S6.
Figure 7. Relative contributions of CBX7 interactions with H3K27me and RNA to senescence regulation
a, Comparison of transcript levels relative to HPRT of ANRIL and INK4a in IMR90 cells undergoing replicative senescence. b, MEFs were infected with the indicated vectors and after selection, growth curves were performed. c, Colony formation assay of MEFs infected with the indicated vectors. Cells were seeded at passage 4. d, Western blots assessing expression levels of p15Ink4b, p16Ink4a and p19Arf in MEFs infected with the indicated constructs. e, Quantitative RT-PCR showing levels of Ink4b, Ink4a and Arf in same cells as in d. f, Model of ncRNA-mediated gene repression of the INK4b/ARF/INK4a locus by the Polycomb group complexes. Schematic model illustrating molecular interplay between methylated H3K27 (by EZH2 of PRC2) and ncRNA transcripts ANRIL for silencing of the INK4b/ARF/INK4a locus by Polycomb CBX7. Both PRC1 and PRC2 complexes are retained at a repression site of a target gene through their association with the nascent ANRIL transcripts of Pol II. Note that the stem loop RNA structure is used for illustration only; RNA/CBX7 binding could result in dissociation of PRC1 from H3K27me, leading to a reversal of transcriptional repression of the target gene. See also Figure S7.
Similar articles
- Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression.
Aguilo F, Zhou MM, Walsh MJ. Aguilo F, et al. Cancer Res. 2011 Aug 15;71(16):5365-9. doi: 10.1158/0008-5472.CAN-10-4379. Epub 2011 Aug 9. Cancer Res. 2011. PMID: 21828241 Free PMC article. Review. - Polycomb mediated epigenetic silencing and replication timing at the INK4a/ARF locus during senescence.
Agherbi H, Gaussmann-Wenger A, Verthuy C, Chasson L, Serrano M, Djabali M. Agherbi H, et al. PLoS One. 2009 May 20;4(5):e5622. doi: 10.1371/journal.pone.0005622. PLoS One. 2009. PMID: 19462008 Free PMC article. - Epigenetic regulation of the INK4b-ARF-INK4a locus: in sickness and in health.
Popov N, Gil J. Popov N, et al. Epigenetics. 2010 Nov-Dec;5(8):685-90. doi: 10.4161/epi.5.8.12996. Epub 2010 Nov 1. Epigenetics. 2010. PMID: 20716961 Free PMC article. Review. - Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene.
Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Kotake Y, et al. Oncogene. 2011 Apr 21;30(16):1956-62. doi: 10.1038/onc.2010.568. Epub 2010 Dec 13. Oncogene. 2011. PMID: 21151178 Free PMC article. - SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus.
Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. Kia SK, et al. Mol Cell Biol. 2008 May;28(10):3457-64. doi: 10.1128/MCB.02019-07. Epub 2008 Mar 10. Mol Cell Biol. 2008. PMID: 18332116 Free PMC article.
Cited by
- LINC00520 is induced by Src, STAT3, and PI3K and plays a functional role in breast cancer.
Henry WS, Hendrickson DG, Beca F, Glass B, Lindahl-Allen M, He L, Ji Z, Struhl K, Beck AH, Rinn JL, Toker A. Henry WS, et al. Oncotarget. 2016 Dec 13;7(50):81981-81994. doi: 10.18632/oncotarget.11962. Oncotarget. 2016. PMID: 27626181 Free PMC article. - Epstein-Barr virus nuclear antigen 3A partially coincides with EBNA3C genome-wide and is tethered to DNA through BATF complexes.
Schmidt SC, Jiang S, Zhou H, Willox B, Holthaus AM, Kharchenko PV, Johannsen EC, Kieff E, Zhao B. Schmidt SC, et al. Proc Natl Acad Sci U S A. 2015 Jan 13;112(2):554-9. doi: 10.1073/pnas.1422580112. Epub 2014 Dec 24. Proc Natl Acad Sci U S A. 2015. PMID: 25540416 Free PMC article. - Crosstalk among Epigenetic Pathways Regulates Neurogenesis.
Jobe EM, McQuate AL, Zhao X. Jobe EM, et al. Front Neurosci. 2012 May 8;6:59. doi: 10.3389/fnins.2012.00059. eCollection 2012. Front Neurosci. 2012. PMID: 22586361 Free PMC article. - A central role for long non-coding RNA in cancer.
Mitra SA, Mitra AP, Triche TJ. Mitra SA, et al. Front Genet. 2012 Feb 15;3:17. doi: 10.3389/fgene.2012.00017. eCollection 2012. Front Genet. 2012. PMID: 22363342 Free PMC article. - A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells.
Johnsson P, Ackley A, Vidarsdottir L, Lui WO, Corcoran M, Grandér D, Morris KV. Johnsson P, et al. Nat Struct Mol Biol. 2013 Apr;20(4):440-6. doi: 10.1038/nsmb.2516. Epub 2013 Feb 24. Nat Struct Mol Biol. 2013. PMID: 23435381 Free PMC article.
References
- Bernard D, Martinez-Leal J, Rizzo S, Martinez D, Hudson D, Visakorpi T, Peters G, Carnero A, Beach D, Gil J. CBX7 controls the growth of normal and tumor-derived prostate cells by repressing the Ink4a/Arf locus. Oncogene. 2005;24:5543–5551. - PubMed
- Bernstein E, Allis C. RNA meets chromatin. Genes Dev. 2005;19:1635–1655. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM073207/GM/NIGMS NIH HHS/United States
- R01 CA087658/CA/NCI NIH HHS/United States
- R01 HG004508/HG/NHGRI NIH HHS/United States
- MC_U120085810/MRC_/Medical Research Council/United Kingdom
- RC1 DA028776/DA/NIDA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases