Structure of the human mTOR complex I and its implications for rapamycin inhibition - PubMed (original) (raw)
Structure of the human mTOR complex I and its implications for rapamycin inhibition
Calvin K Yip et al. Mol Cell. 2010.
Abstract
The mammalian target of rapamycin complex 1 (mTORC1) regulates cell growth in response to the nutrient and energy status of the cell, and its deregulation is common in human cancers. Little is known about the overall architecture and subunit organization of this essential signaling complex. We have determined the three-dimensional (3D) structure of the fully assembled human mTORC1 by cryo-electron microscopy (cryo-EM). Our analyses reveal that mTORC1 is an obligate dimer with an overall rhomboid shape and a central cavity. The dimeric interfaces are formed by interlocking interactions between the mTOR and raptor subunits. Extended incubation with FKBP12-rapamycin compromises the structural integrity of mTORC1 in a stepwise manner, leading us to propose a model in which rapamycin inhibits mTORC1-mediated phosphorylation of 4E-BP1 and S6K1 through different mechanisms.
Copyright (c) 2010 Elsevier Inc. All rights reserved.
Figures
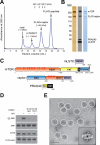
Figure 1
mTORC1 purification. (A) mTORC1 and FLAG-raptor were purified by tandem gel filtration chromatography. Their masses were estimated based on known molecular weight standards as indicated (Thyroglobulin (669 kDa), Ferritin (440 kDa), Aldolase (158 kDa) and Conalbumin (75 kDa)). (B) The gel filtration fraction corresponding to mTORC1 was analyzed by SDS-PAGE followed by silver and Coomassie staining as well as immunoblotting for indicated proteins. (C) Schematics of mTORC1 components illustrating the various predicted domains. (D) In vitro kinase assay showing that purified mTORC1 phosphorylates S6K1 and is inhibited by both rapamycin-FKBP12 (rapa) and Torin1. (E) EM of negatively stained mTORC1. A raw image of mTORC1 particles (circled) and a representative class average from the classification of 10,080 particles (inset). The scale bar represents 50 nm, and the side length of the panel showing the class average is 45 nm. “see also Figure S1”
Figure 2
Cryo-EM reconstruction of mTORC1 and its molecular organization. (A) Image of a vitrified specimen showing individual mTORC1 particles (circled). The scale bar represents 50 nm. (B) Different views of the 3D reconstruction of mTORC1 filtered to 26 Å, with the main structural features denoted. The scale bar represents 5 nm. (C) Molecular organization of mTORC1. Left panels: Representative class averages from antibody labeling experiments of mTORC1 (top) and schematic representations showing mTORC1 in white and the antibody in pale red (bottom). The side length of each panel is 45 nm. Right panel: Location of raptor, mLST8, and PRAS40 in the cryo-EM density map of mTORC1. “see also Figure S2”
Figure 3
3D reconstruction of raptor and molecular docking. (A) Silver-stained gel and immunoblot of the gel filtration fraction containing free raptor detected the presence of PRAS40. (B) EM image of negatively stained raptor (circled) and two representative class averages from the classification of 12,216 particles (bottom right insets). Class II particles contain an additional density (red arrow) compared to Class I particles, which likely represents PRAS40. The scale bar represents 25 nm, and the side length of the panels showing the class averages is 27 nm. (C) Different views of the raptor 3D reconstruction. The scale bar represents 2.5 nm. (D) Two copies of the raptor 3D reconstruction (gold) and two models of a representative WD40 domain (PDB code 3EMH, red) were placed into the cryo-EM density map (gray). The green asterisk depicts the location of PRAS40 as determined by antibody labeling. The blue dotted line represents the dimer interface. (E) The proposed locations of the N- and C-terminal domains (marked “N” and “C”) and the kinase domain of mTOR (purple star). The black lines labeled “I” and “II” delineate the two interaction interfaces formed by each mTOR molecule with the two raptor subunits. “see also Figure S3”
Figure 4
Effects of rapamycin-FKBP12 on mTORC1. (A) Representative class averages of untreated mTORC1 (left) and mTORC1 treated with 50 nM of rapamycin and 0.02 μg/μl GST-FKBP12 (middle), and a schematic representation showing the additional density in pale red (right). The side length of each panel is 45 nm. To the right, the location of the FRB domain with respect to the other components in the cryo-EM map of mTORC1 is shown. (B) and (C) Purified mTORC1 was treated with 50 nM of rapamycin and 0.02 μg/μl GST-FKBP12 or 100 nM Torin1. EM images of negatively stained samples were taken at the indicated time points. The inset in each image shows an enlarged view of the area marked by the white square. The scale bars represent 100 nm. (D) and (E) mTOR immunoprecipitates, prepared in lysis buffers containing 0.3% CHAPS or 1% Triton X-100, were subjected to in vitro kinase assays using 4E-BP1 or S6K1 as a substrate in the presence of 100 nM rapamycin and 0.02 μg/μl FKBP12 or 100 nM Torin1. Assays were then analyzed by immunoblotting for the indicated proteins and phosphorylation states. (F) A model depicting a potential mechanism of mTORC1 inhibition by FKBP12-rapamycin. “see also Figure S4”
Similar articles
- PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding.
Wang L, Harris TE, Roth RA, Lawrence JC Jr. Wang L, et al. J Biol Chem. 2007 Jul 6;282(27):20036-44. doi: 10.1074/jbc.M702376200. Epub 2007 May 17. J Biol Chem. 2007. PMID: 17510057 - Rapamycin inhibits cytoskeleton reorganization and cell motility by suppressing RhoA expression and activity.
Liu L, Luo Y, Chen L, Shen T, Xu B, Chen W, Zhou H, Han X, Huang S. Liu L, et al. J Biol Chem. 2010 Dec 3;285(49):38362-73. doi: 10.1074/jbc.M110.141168. Epub 2010 Oct 11. J Biol Chem. 2010. PMID: 20937815 Free PMC article. - Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40.
Yang H, Jiang X, Li B, Yang HJ, Miller M, Yang A, Dhar A, Pavletich NP. Yang H, et al. Nature. 2017 Dec 21;552(7685):368-373. doi: 10.1038/nature25023. Epub 2017 Dec 13. Nature. 2017. PMID: 29236692 Free PMC article. - LKB1 and AMP-activated protein kinase control of mTOR signalling and growth.
Shaw RJ. Shaw RJ. Acta Physiol (Oxf). 2009 May;196(1):65-80. doi: 10.1111/j.1748-1716.2009.01972.x. Epub 2009 Feb 19. Acta Physiol (Oxf). 2009. PMID: 19245654 Free PMC article. Review. - Not all substrates are treated equally: implications for mTOR, rapamycin-resistance and cancer therapy.
Choo AY, Blenis J. Choo AY, et al. Cell Cycle. 2009 Feb 15;8(4):567-72. doi: 10.4161/cc.8.4.7659. Epub 2009 Feb 18. Cell Cycle. 2009. PMID: 19197153 Review.
Cited by
- mTOR kinase: a possible pharmacological target in the management of chronic pain.
Lisi L, Aceto P, Navarra P, Dello Russo C. Lisi L, et al. Biomed Res Int. 2015;2015:394257. doi: 10.1155/2015/394257. Epub 2015 Jan 1. Biomed Res Int. 2015. PMID: 25685786 Free PMC article. Review. - Analysis of PI3K/mTOR Pathway Biomarkers and Their Prognostic Value in Women with Hormone Receptor-Positive, HER2-Negative Early Breast Cancer.
Azim HA, Kassem L, Treilleux I, Wang Q, El Enein MA, Anis SE, Bachelot T. Azim HA, et al. Transl Oncol. 2016 Apr;9(2):114-123. doi: 10.1016/j.tranon.2016.01.001. Transl Oncol. 2016. PMID: 27084427 Free PMC article. - Lysosomes at the Crossroads of Cell Metabolism, Cell Cycle, and Stemness.
Nowosad A, Besson A. Nowosad A, et al. Int J Mol Sci. 2022 Feb 18;23(4):2290. doi: 10.3390/ijms23042290. Int J Mol Sci. 2022. PMID: 35216401 Free PMC article. Review. - Cryo-EM insight into the structure of MTOR complex 1 and its interactions with Rheb and substrates.
Chao LH, Avruch J. Chao LH, et al. F1000Res. 2019 Jan 3;8:F1000 Faculty Rev-14. doi: 10.12688/f1000research.16109.1. eCollection 2019. F1000Res. 2019. PMID: 30647914 Free PMC article. Review. - The role of raptor in the mechanical load-induced regulation of mTOR signaling, protein synthesis, and skeletal muscle hypertrophy.
You JS, McNally RM, Jacobs BL, Privett RE, Gundermann DM, Lin KH, Steinert ND, Goodman CA, Hornberger TA. You JS, et al. FASEB J. 2019 Mar;33(3):4021-4034. doi: 10.1096/fj.201801653RR. Epub 2018 Dec 3. FASEB J. 2019. PMID: 30509128 Free PMC article.
References
- Adami A, Garcia-Alvarez B, Arias-Palomo E, Barford D, Llorca O. Structure of TOR and its complex with KOG1. Mol Cell. 2007;27:509–516. - PubMed
- Choi JW, Chen J, Schreiber SL, Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239–242. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CAPMC/ CIHR/Canada
- CA103866/CA/NCI NIH HHS/United States
- R01 CA129105/CA/NCI NIH HHS/United States
- AI47389/AI/NIAID NIH HHS/United States
- R01 AI047389/AI/NIAID NIH HHS/United States
- R01 CA103866/CA/NCI NIH HHS/United States
- R37 AI047389/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous