Switch from stress response to homeobox transcription factors in adipose tissue after profound fat loss - PubMed (original) (raw)
. 2010 Jun 9;5(6):e11033.
doi: 10.1371/journal.pone.0011033.
Dag J Fadnes, Anne-Kristin Stavrum, Christine Stansberg, Rita Holdhus, Tuyen Hoang, Vivian L Veum, Bjørn Jostein Christensen, Villy Våge, Jørn V Sagen, Vidar M Steen, Gunnar Mellgren
Affiliations
- PMID: 20543949
- PMCID: PMC2882947
- DOI: 10.1371/journal.pone.0011033
Switch from stress response to homeobox transcription factors in adipose tissue after profound fat loss
Simon N Dankel et al. PLoS One. 2010.
Abstract
Background: In obesity, impaired adipose tissue function may promote secondary disease through ectopic lipid accumulation and excess release of adipokines, resulting in systemic low-grade inflammation, insulin resistance and organ dysfunction. However, several of the genes regulating adipose tissue function in obesity are yet to be identified.
Methodology/principal findings: In order to identify novel candidate genes that may regulate adipose tissue function, we analyzed global gene expression in abdominal subcutaneous adipose tissue before and one year after bariatric surgery (biliopancreatic diversion with duodenal switch, BPD/DS) (n = 16). Adipose tissue from lean healthy individuals was also analyzed (n = 13). Two different microarray platforms (AB 1700 and Illumina) were used to measure the differential gene expression, and the results were further validated by qPCR. Surgery reduced BMI from 53.3 to 33.1 kg/m(2). The majority of differentially expressed genes were down-regulated after profound fat loss, including transcription factors involved in stress response, inflammation, and immune cell function (e.g., FOS, JUN, ETS, C/EBPB, C/EBPD). Interestingly, a distinct set of genes was up-regulated after fat loss, including homeobox transcription factors (IRX3, IRX5, HOXA5, HOXA9, HOXB5, HOXC6, EMX2, PRRX1) and extracellular matrix structural proteins (COL1A1, COL1A2, COL3A1, COL5A1, COL6A3).
Conclusions/significance: The data demonstrate a marked switch of transcription factors in adipose tissue after profound fat loss, providing new molecular insight into a dichotomy between stress response and metabolically favorable tissue development. Our findings implicate homeobox transcription factors as important regulators of adipose tissue function.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
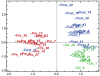
Figure 1. Correspondence Analysis showing projection of samples before and after bariatric surgery and lean healthy controls.
The first principal component (10.69% component variance) separated the pre-operative samples from the post-operative and control samples, and the second principal component (6.36% component variance) separated the post-operative from control samples. The post-operative sample of patient 17 was similar to the pre-operative samples, suggesting an overall unaltered gene expression pattern after surgery for this patient. A technical error may have occurred, but we cannot rule out a unique gene expression pattern in the adipose tissue of this patient.
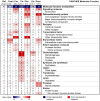
Figure 2. Functional categorization of differentially expressed genes in subcutaneous adipose tissue after fat loss (Biological Process).
PANTHER was used to search for over-represented Biological Process categories among the most differentially expressed genes (q-value = 0, fold change ≥1.5), comparing adipose tissue before versus after bariatric surgery (n = 16) and versus controls (n = 13). The color intensity displays the statistical significance (−log p-value) of over- and under-represented PANTHER functional categories. A p-value<0.01 with Bonferroni correction for multiple testing was used as inclusion criterion for categories. Numbers presented in the table indicate the percentage of genes within a gene set that map to the given category, e.g. 18% of the 469 down-regulated genes map to the biological process ‘Immunity and defense’. The first column states the overall distribution of a term among all human NCBI genes (25,431), e.g. 5% of the genes are expected to map to ‘Immunity and defense’, hence this category is significantly over-represented among the down-regulated genes. Ref, reference (based on all human NCBI genes); Pre, pre-surgery biopsies; Post, post-surgery biopsies; Ctr, biopsies of lean controls; Arrow up, up-regulated/more expressed genes; Arrow down, down-regulated/less expressed genes.
Figure 3. Functional categorization of differentially expressed genes in subcutaneous adipose tissue after fat loss (Molecular Function).
PANTHER was used to search for over-represented Molecular Function categories among the most differentially expressed genes (q-value = 0, fold change ≥1.5), comparing adipose tissue before versus after bariatric surgery (n = 16) and versus controls (n = 13). The color intensity displays the statistical significance (−log p-value) of over- and under-represented PANTHER functional categories. A p-value<0.01 with Bonferroni correction for multiple testing was used as inclusion criterion for categories. Numbers presented in the table indicate the percentage of genes within a gene set that map to the given category, e.g. 8% of the 469 down-regulated genes map to the molecular function ‘Signalling molecule’. The first column states the overall distribution of a term among all human NCBI genes (25,431), e.g. 3% of the genes are expected to map to ‘Signalling molecule’, hence this category is significantly over-represented among the down-regulated genes. Ref, reference (based on all human NCBI genes); Pre, pre-surgery biopsies; Post, post-surgery biopsies; Ctr, biopsies of lean controls; Arrow up, up-regulated/more expressed genes; Arrow down, down-regulated/less expressed genes.
Similar articles
- COL6A3 expression in adipose tissue cells is associated with levels of the homeobox transcription factor PRRX1.
Dankel SN, Grytten E, Bjune JI, Nielsen HJ, Dietrich A, Blüher M, Sagen JV, Mellgren G. Dankel SN, et al. Sci Rep. 2020 Nov 19;10(1):20164. doi: 10.1038/s41598-020-77406-2. Sci Rep. 2020. PMID: 33214660 Free PMC article. - The nuclear receptors NUR77, NURR1 and NOR1 in obesity and during fat loss.
Veum VL, Dankel SN, Gjerde J, Nielsen HJ, Solsvik MH, Haugen C, Christensen BJ, Hoang T, Fadnes DJ, Busch C, Våge V, Sagen JV, Mellgren G. Veum VL, et al. Int J Obes (Lond). 2012 Sep;36(9):1195-202. doi: 10.1038/ijo.2011.240. Epub 2011 Dec 6. Int J Obes (Lond). 2012. PMID: 22143616 - Hoxa5 undergoes dynamic DNA methylation and transcriptional repression in the adipose tissue of mice exposed to high-fat diet.
Parrillo L, Costa V, Raciti GA, Longo M, Spinelli R, Esposito R, Nigro C, Vastolo V, Desiderio A, Zatterale F, Ciccodicola A, Formisano P, Miele C, Beguinot F. Parrillo L, et al. Int J Obes (Lond). 2016 Jun;40(6):929-37. doi: 10.1038/ijo.2016.36. Epub 2016 Mar 16. Int J Obes (Lond). 2016. PMID: 26980478 - Adipose Tissue Composition in Obesity and After Bariatric Surgery.
Adami GF, Carbone F, Montecucco F, Camerini G, Cordera R. Adami GF, et al. Obes Surg. 2019 Sep;29(9):3030-3038. doi: 10.1007/s11695-019-04030-z. Obes Surg. 2019. PMID: 31190263 Review. - The Transcription Factor HOXA5: Novel Insights into Metabolic Diseases and Adipose Tissue Dysfunction.
Parrillo L, Spinelli R, Longo M, Zatterale F, Santamaria G, Leone A, Campitelli M, Raciti GA, Beguinot F. Parrillo L, et al. Cells. 2023 Aug 18;12(16):2090. doi: 10.3390/cells12162090. Cells. 2023. PMID: 37626900 Free PMC article. Review.
Cited by
- Nutritionally mediated oxidative stress and inflammation.
Muñoz A, Costa M. Muñoz A, et al. Oxid Med Cell Longev. 2013;2013:610950. doi: 10.1155/2013/610950. Epub 2013 Jun 13. Oxid Med Cell Longev. 2013. PMID: 23844276 Free PMC article. Review. - Inflammation triggers specific microRNA profiles in human adipocytes and macrophages and in their supernatants.
Ortega FJ, Moreno M, Mercader JM, Moreno-Navarrete JM, Fuentes-Batllevell N, Sabater M, Ricart W, Fernández-Real JM. Ortega FJ, et al. Clin Epigenetics. 2015 Apr 24;7(1):49. doi: 10.1186/s13148-015-0083-3. eCollection 2015. Clin Epigenetics. 2015. PMID: 25926893 Free PMC article. - Weighted Gene Co-Expression Network Analysis Identifies Key Modules and Central Genes Associated With Bovine Subcutaneous Adipose Tissue.
Sheng H, Pan C, Wang S, Yang C, Zhang J, Hu C, Hu H, Feng X, Yang M, Lei Z, Gao Y, Wang Z, Ma Y. Sheng H, et al. Front Vet Sci. 2022 Jun 22;9:914848. doi: 10.3389/fvets.2022.914848. eCollection 2022. Front Vet Sci. 2022. PMID: 35812879 Free PMC article. - Paired related homeobox 1 attenuates autophagy via acetyl-CoA carboxylase 1-regulated fatty acid metabolism in salivary adenoid cystic carcinoma.
Gao J, Ma K, Zhang L, Li T, Zhao B, Jiang Y. Gao J, et al. FEBS Open Bio. 2022 May;12(5):1006-1016. doi: 10.1002/2211-5463.13367. Epub 2022 Mar 29. FEBS Open Bio. 2022. PMID: 35032368 Free PMC article. - Weight loss normalizes enhanced expression of the oncogene survivin in visceral adipose tissue and blood leukocytes from individuals with obesity.
Izquierdo AG, Carreira MC, Rodriguez-Carnero G, Fernandez-Quintela A, Sueiro AM, Martinez-Olmos MA, Guzman G, De Luis D, Pinhel MAS, Nicoletti CF, Nonino CB, Ortega FJ, Portillo MP, Fernandez-Real JM, Casanueva FF, Crujeiras AB. Izquierdo AG, et al. Int J Obes (Lond). 2021 Jan;45(1):206-216. doi: 10.1038/s41366-020-0630-7. Epub 2020 Jun 16. Int J Obes (Lond). 2021. PMID: 32546857 Free PMC article.
References
- Poulos SP, Hausman DB, Hausman GJ. The development and endocrine functions of adipose tissue. Mol Cell Endocrinol 2009 - PubMed
- Bays HE, Gonzalez-Campoy JM, Bray GA, Kitabchi AE, Bergman DA, et al. Pathogenic potential of adipose tissue and metabolic consequences of adipocyte hypertrophy and increased visceral adiposity. Expert Rev Cardiovasc Ther. 2008;6:343–368. - PubMed
- Bluher M. Adipose tissue dysfunction in obesity. Exp Clin Endocrinol Diabetes. 2009;117:241–250. - PubMed
- Spalding KL, Arner E, Westermark PO, Bernard S, Buchholz BA, et al. Dynamics of fat cell turnover in humans. Nature. 2008;453:783–787. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous