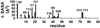Determinants of affinity and activity of the anti-sigma factor AsiA - PubMed (original) (raw)
Determinants of affinity and activity of the anti-sigma factor AsiA
Joshua M Gilmore et al. Biochemistry. 2010.
Abstract
The AsiA protein is a T4 bacteriophage early gene product that regulates transcription of host and viral genes. Monomeric AsiA binds tightly to the sigma(70) subunit of Escherichia coli RNA polymerase, thereby inhibiting transcription from bacterial promoters and phage early promoters and coactivating transcription from phage middle promoters. Results of structural studies have identified amino acids at the protomer-protomer interface in dimeric AsiA and at the monomeric AsiA-sigma(70) interface and demonstrated substantial overlap in the sets of residues that comprise each. Here we evaluate the contributions of individual interfacial amino acid side chains to protomer-protomer affinity in AsiA homodimers, to monomeric AsiA affinity for sigma(70), and to AsiA function in transcription. Sedimentation equilibrium, dynamic light scattering, electrophoretic mobility shift, and transcription activity measurements were used to assess affinity and function of site-specific AsiA mutants. Alanine substitutions for solvent-inaccessible residues positioned centrally in the protomer-protomer interface of the AsiA homodimer, V14, I17, and I40, resulted in the largest changes in free energy of dimer association, whereas alanine substitutions at other interfacial positions had little effect. These residues also contribute significantly to AsiA-dependent regulation of RNA polymerase activity, as do additional residues positioned at the periphery of the interface (K20 and F21). Notably, the relative contributions of a given amino acid side chain to RNA polymerase inhibition and activation (MotA-independent) by AsiA are very similar in most cases. The mainstay for intermolecular affinity and AsiA function appears to be I17. Our results define the core interfacial residues of AsiA, establish roles for many of the interfacial amino acids, are in agreement with the tenets underlying protein-protein interactions and interfaces, and will be beneficial for a general, comprehensive understanding of the mechanistic underpinnings of bacterial RNA polymerase regulation.
Figures
FIGURE 1
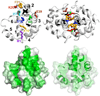
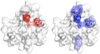
The dimer interface of AsiA. (Top) Site directed amino acid substitutions in AsiA. Eleven Ala mutants of AsiA were produced to evaluate the functions of the replaced side chains. These side chains are shown on ribbon models of monomeric AsiA (left) and dimeric AsiA (right). Most of these are at the homodimer interface and include M1 (purple), E10 (gold), V14 (blue), I17 (cyan), K20 (red), F21 (orange), E39 (brown), and I40 (green). The double mutant I17A/I40A (magenta) was also produced. Unstable proteins resulted from mutation of the amino acids with side chains colored black (L18, V27, V42) to Ala. The six helices of AsiA are numbered from the N-terminus. (Bottom) The hydrophobic character of the interface region of AsiA. The hydropathies of the individual residues are plotted on the Connolly surface of one protomer of dimeric AsiA, with green representing the most hydrophobic and white the least. The large hydrophobic cleft in AsiA is apparent, formed by the side chains of residues from helices 1–3 including V14, I17, L18, V27, F21, I40, and V42. The dimer interface is constructed by aligning the C-terminal end of helix 1 of each protomer with the hydrophobic cleft of the other.
FIGURE 2
AsiA and mutant AsiA binding to σ70. Electrophoretic mobility shift assays using non-denaturing PAGE were used to evaluate the apparent affinities of AsiA and the mutant AsiA proteins for σ70 by monitoring formation of the AsiA-σ70 complexes. Non-denaturing PAGE was also used to confirm the purities of the AsiA proteins and to demonstrate their relative electrophoretic mobilities, as shown in the first (upper left) panel. The remaining panels show the results when increasing concentrations of AsiA or mutant AsiA were incubated with a fixed concentration of σ70. Electrophoresis of the samples was performed as described in the text. Lane 1, AsiA or mutant AsiA only (20 µM). Lane 2, σ70 only (20 µM). Lanes 3–7, 20 µM σ70 and 5, 10, 20, 40, or 80 µM AsiA or mutant AsiA, respectively.
FIGURE 3
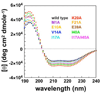
Circular dichroism spectra for AsiA and the AsiA mutants. The far-UV circular dichroism spectra of the mutant proteins are similar to one another and to wild type and clearly indicate no loss of helical secondary structure in the mutants. Shown are spectra for wild type AsiA (black) and mutants M1A (purple), E10A (gold), V14A (blue), I17A (cyan), K20A (red), F21A (orange), E39A (brown), I40A (green), and the double mutant I17A/I40A (magenta).
FIGURE 4
NMR spectra of the AsiA mutants. The 1H, 15N-HSQC spectra of wild type AsiA and the stable mutant AsiA proteins are shown. Although some slight (K20A) to moderate (I17A) conformational averaging is observed, the chemical shifts and chemical shift dispersion observed in the spectra of the mutants are similar to wild type and indicate that the mutations do not eliminate tertiary structure.
FIGURE 5
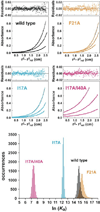
Sedimentation equilibrium results for AsiA and AsiA mutant proteins. (Top) For each protein, sedimentation equilibrium data were acquired at three protein concentrations and three centrifugation speeds. Shown are representative results at 40,000 rpm for 3.0, 7.0, and 12.0 µM protein (wild type, I17A, F21A, and the I17A/I40A double mutant) monitored at 230 nm. In each case, the best fit from the global analysis (shown as a black line through the data points) was to a monomer-dimer equilibrium model. Residuals are shown at the top of each panel. (Bottom) Distributions of values for ln _K_a from Monte Carlo simulations for wild type AsiA and the I17A, F21A, and I17A/I40A mutants.
FIGURE 6
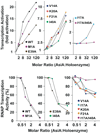
Transcription inhibition and activation by AsiA and mutant AsiA proteins. The ability of AsiA and mutant AsiA proteins to inhibit or activate transcription by E. coli RNA polymerase are shown. (Top) Activation of transcription from the T7 bacteriophage early promoter A1. (Bottom) Inhibition of transcription from the T4 bacteriophage PrIIB2 middle promoter.
FIGURE 7
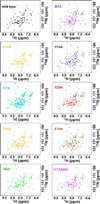
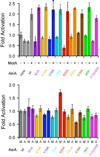
MotA-dependent transcription co-activation by AsiA and mutant AsiA proteins. (Top) Fold activation with or without MotA in the presence of saturating wild type AsiA or mutant AsiA, normalized to the activity in the absence of both MotA and AsiA. (Bottom) Fold activation by MotA or wild type AsiA and AsiA mutants normalized to wild-type activation. “M” on the abscissa represents activation by MotA in the presence of saturating wild type AsiA or mutant AsiA, with activation normalized to wild type AsiA. “A” on the abscissa represents activation by wild type AsiA (or mutant AsiA) in the presence of MotA, normalized to wild type AsiA. Error limits are ± 1 standard deviation.
FIGURE 8
Solvent accessible surface area differences between AsiA dimers and protomers. The differences shown are residue localized increases in non-polar solvent accessible surface areas for AsiA protomers (from the dimer structures, but in the absence of the opposing protomer) relative to dimers. The protomer-protomer interface in dimeric AsiA is approximately 2012 ± 102 Å2 (1253 ± 102 Å2 of this is non-polar), with 1741 ± 74 Å2 contributed by amino acid side chains (error limits are standard deviations computed from the ensemble of 25 deposited NMR structures).
FIGURE 9
Functional epitopes of AsiA maintaining (left) protomer-protomer affinity in dimeric AsiA and (right) AsiA-σ70 affinity. Color intensity is a qualitative indicator of relative contribution.
Similar articles
- Mapping the molecular interface between the sigma(70) subunit of E. coli RNA polymerase and T4 AsiA.
Minakhin L, Camarero JA, Holford M, Parker C, Muir TW, Severinov K. Minakhin L, et al. J Mol Biol. 2001 Mar 2;306(4):631-42. doi: 10.1006/jmbi.2001.4445. J Mol Biol. 2001. PMID: 11243776 - Interaction of T4 AsiA with its target sites in the RNA polymerase sigma70 subunit leads to distinct and opposite effects on transcription.
Minakhin L, Niedziela-Majka A, Kuznedelov K, Adelman K, Urbauer JL, Heyduk T, Severinov K. Minakhin L, et al. J Mol Biol. 2003 Feb 21;326(3):679-90. doi: 10.1016/s0022-2836(02)01442-0. J Mol Biol. 2003. PMID: 12581632 - Transcriptional takeover by sigma appropriation: remodelling of the sigma70 subunit of Escherichia coli RNA polymerase by the bacteriophage T4 activator MotA and co-activator AsiA.
Hinton DM, Pande S, Wais N, Johnson XB, Vuthoori M, Makela A, Hook-Barnard I. Hinton DM, et al. Microbiology (Reading). 2005 Jun;151(Pt 6):1729-1740. doi: 10.1099/mic.0.27972-0. Microbiology (Reading). 2005. PMID: 15941982 Review. - Transcription regulation by bacteriophage T4 AsiA.
Minakhin L, Severinov K. Minakhin L, et al. Protein Expr Purif. 2005 May;41(1):1-8. doi: 10.1016/j.pep.2004.09.019. Protein Expr Purif. 2005. PMID: 15802215 Review.
Cited by
- Identification of a phosphorylation site within the P protein important for mRNA transcription and growth of parainfluenza virus 5.
Sun D, Luthra P, Xu P, Yoon H, He B. Sun D, et al. J Virol. 2011 Aug;85(16):8376-85. doi: 10.1128/JVI.00618-11. Epub 2011 Jun 15. J Virol. 2011. PMID: 21680523 Free PMC article. - Environmental T4-Family Bacteriophages Evolve to Escape Abortive Infection via Multiple Routes in a Bacterial Host Employing "Altruistic Suicide" through Type III Toxin-Antitoxin Systems.
Chen B, Akusobi C, Fang X, Salmond GPC. Chen B, et al. Front Microbiol. 2017 May 31;8:1006. doi: 10.3389/fmicb.2017.01006. eCollection 2017. Front Microbiol. 2017. PMID: 28620370 Free PMC article. - Visualizing the phage T4 activated transcription complex of DNA and E. coli RNA polymerase.
James TD, Cardozo T, Abell LE, Hsieh ML, Jenkins LM, Jha SS, Hinton DM. James TD, et al. Nucleic Acids Res. 2016 Sep 19;44(16):7974-88. doi: 10.1093/nar/gkw656. Epub 2016 Jul 25. Nucleic Acids Res. 2016. PMID: 27458207 Free PMC article. - Solution structure and properties of AlgH from Pseudomonas aeruginosa.
Urbauer JL, Cowley AB, Broussard HP, Niedermaier HT, Bieber Urbauer RJ. Urbauer JL, et al. Proteins. 2015 Jun;83(6):1137-50. doi: 10.1002/prot.24811. Epub 2015 Apr 29. Proteins. 2015. PMID: 25857636 Free PMC article. - Mycobacterium tuberculosis RNA polymerase-binding protein A (RbpA) and its interactions with sigma factors.
Bortoluzzi A, Muskett FW, Waters LC, Addis PW, Rieck B, Munder T, Schleier S, Forti F, Ghisotti D, Carr MD, O'Hare HM. Bortoluzzi A, et al. J Biol Chem. 2013 May 17;288(20):14438-14450. doi: 10.1074/jbc.M113.459883. Epub 2013 Apr 2. J Biol Chem. 2013. PMID: 23548911 Free PMC article.
References
- Stevens A. Inhibition of DNA-enzyme binding by an RNA polymerase inhibitor from T4 phage-infected Escherichia coli. Biochim Biophys Acta. 1977;475:193–196. - PubMed
- Stevens A, Rhoton JC. Characterization of an inhibitor causing potassium chloride sensitivity of an RNA polymerase from T4 phage-infected Escherichia coli. Biochemistry. 1975;14:5074–5079. - PubMed
- Stevens A. Deoxyribonucleic acid dependent ribonucleic acid polymerases from two T4 phage-infected systems. Biochemistry. 1974;13:493–503. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM059295-05/GM/NIGMS NIH HHS/United States
- R01 GM054998-05/GM/NIGMS NIH HHS/United States
- R01 GM059295-02/GM/NIGMS NIH HHS/United States
- GM54998/GM/NIGMS NIH HHS/United States
- R01 GM059295-06/GM/NIGMS NIH HHS/United States
- R01 GM059295-09/GM/NIGMS NIH HHS/United States
- R01 GM054998-07/GM/NIGMS NIH HHS/United States
- R01 GM059295-08/GM/NIGMS NIH HHS/United States
- R01 GM054998-04/GM/NIGMS NIH HHS/United States
- R01 GM059295-04/GM/NIGMS NIH HHS/United States
- R01 GM054998-10/GM/NIGMS NIH HHS/United States
- R01 GM054998-08/GM/NIGMS NIH HHS/United States
- R01 GM054998-11/GM/NIGMS NIH HHS/United States
- R01 GM059295-01/GM/NIGMS NIH HHS/United States
- R01 GM059295/GM/NIGMS NIH HHS/United States
- GM59295/GM/NIGMS NIH HHS/United States
- R01 GM054998-02/GM/NIGMS NIH HHS/United States
- R01 GM054998-06/GM/NIGMS NIH HHS/United States
- R01 GM054998/GM/NIGMS NIH HHS/United States
- R01 GM059295-03/GM/NIGMS NIH HHS/United States
- R01 GM054998-09/GM/NIGMS NIH HHS/United States
- R01 GM054998-03/GM/NIGMS NIH HHS/United States
- R01 GM059295-07/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources