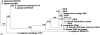Metagenomic detection of phage-encoded platelet-binding factors in the human oral cavity - PubMed (original) (raw)
. 2011 Mar 15;108 Suppl 1(Suppl 1):4547-53.
doi: 10.1073/pnas.1000089107. Epub 2010 Jun 14.
Affiliations
- PMID: 20547834
- PMCID: PMC3063595
- DOI: 10.1073/pnas.1000089107
Metagenomic detection of phage-encoded platelet-binding factors in the human oral cavity
Dana Willner et al. Proc Natl Acad Sci U S A. 2011.
Abstract
The human oropharynx is a reservoir for many potential pathogens, including streptococcal species that cause endocarditis. Although oropharyngeal microbes have been well described, viral communities are essentially uncharacterized. We conducted a metagenomic study to determine the composition of oropharyngeal DNA viral communities (both phage and eukaryotic viruses) in healthy individuals and to evaluate oropharyngeal swabs as a rapid method for viral detection. Viral DNA was extracted from 19 pooled oropharyngeal swabs and sequenced. Viral communities consisted almost exclusively of phage, and complete genomes of several phage were recovered, including Escherichia coli phage T3, Propionibacterium acnes phage PA6, and Streptococcus mitis phage SM1. Phage relative abundances changed dramatically depending on whether samples were chloroform treated or filtered to remove microbial contamination. pblA and pblB genes of phage SM1 were detected in the metagenomes. pblA and pblB mediate the attachment of S. mitis to platelets and play a significant role in S. mitis virulence in the endocardium, but have never previously been detected in the oral cavity. These genes were also identified in salivary metagenomes from three individuals at three time points and in individual saliva samples by PCR. Additionally, we demonstrate that phage SM1 can be induced by commonly ingested substances. Our results indicate that the oral cavity is a reservoir for pblA and pblB genes and for phage SM1 itself. Further studies will determine the association between pblA and pblB genes in the oral cavity and the risk of endocarditis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
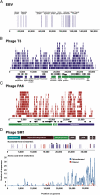
Fig. 1.
Coverage of viral genomes by oropharyngeal metagenomic sequences: Epstein-Barr virus (A), E. coli phage T3 (B), P. acnes phage PA6 (C), and S. mitis phage SM1 (D). Similarities obtained from the chloroformed metagenome are shown in blue, and those from the filtered metagenome are shown in red. Nucleotide-level coverage (A, B, C, and top of D) was determined by alignment of metagenomic sequences to complete viral genome sequences obtained from the National Center for Biotechnology Information using BLAT. Amino acid level coverage (D, bottom) was plotted using significant tBLASTx (_e_-value <10−5) similarities to each genome. Contigs were assembled using the 454 gsAssembler and aligned to genomes using BLAT.
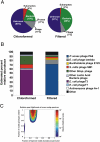
Fig. 2.
Taxonomic composition and diversity of the oropharyngeal metagenomes. (A) Composition of complete metagenomes as determined by best tBLASTx similarities to the nonredundant database (_e_-value <10−5). (_B_) Composition of viral communities. Viral relative abundances were determined by GAAS on the basis of tBLASTx similarities (_e_-value <10−5, percentage identity >30%, query coverage >80%) to a database containing all complete viral genomes currently available at the National Center for Biotechnology Information. (C) Monte Carlo analysis of cross-contig spectra for oropharyngeal metagenomes. The area of maximum likelihood is indicated by an arrow. The metagenomes were predicted to share more than 95% of genotypes with 30% of their relative abundances permuted.
Fig. 3.
Phylogenetic relationships between pblA sequences from saliva samples and reference genomes. The Bayes values show the proportion of sampled trees in which the sequences to the right of the branch point clustered together.
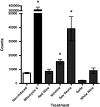
Fig. 4.
Phage induction assay. Data are presented as induction treatment on the x axis versus the mean number of phage events counted using flow cytometry (±SEM) on the y axis (n = 3). White wine, nicotine, soda, and soy sauce treatments were diluted 1:10, and the red wine treatment was diluted 1:100. Asterisks indicate that nicotine, soy sauce, and mitomycin C caused a statistically significant increase in phage count (P < 0.05).
Similar articles
- Proteins PblA and PblB of Streptococcus mitis, which promote binding to human platelets, are encoded within a lysogenic bacteriophage.
Bensing BA, Siboo IR, Sullam PM. Bensing BA, et al. Infect Immun. 2001 Oct;69(10):6186-92. doi: 10.1128/IAI.69.10.6186-6192.2001. Infect Immun. 2001. PMID: 11553559 Free PMC article. - Mechanism of cell surface expression of the Streptococcus mitis platelet binding proteins PblA and PblB.
Mitchell J, Siboo IR, Takamatsu D, Chambers HF, Sullam PM. Mitchell J, et al. Mol Microbiol. 2007 May;64(3):844-57. doi: 10.1111/j.1365-2958.2007.05703.x. Mol Microbiol. 2007. PMID: 17462028 - Genomic organization and molecular characterization of SM1, a temperate bacteriophage of Streptococcus mitis.
Siboo IR, Bensing BA, Sullam PM. Siboo IR, et al. J Bacteriol. 2003 Dec;185(23):6968-75. doi: 10.1128/JB.185.23.6968-6975.2003. J Bacteriol. 2003. PMID: 14617660 Free PMC article. - Bacteriophages infecting Propionibacterium acnes.
Brüggemann H, Lood R. Brüggemann H, et al. Biomed Res Int. 2013;2013:705741. doi: 10.1155/2013/705741. Epub 2013 Apr 11. Biomed Res Int. 2013. PMID: 23691509 Free PMC article. Review. - The novel species Streptococcus tigurinus and its association with oral infection.
Zbinden A, Bostanci N, Belibasakis GN. Zbinden A, et al. Virulence. 2015;6(3):177-82. doi: 10.4161/21505594.2014.970472. Epub 2014 Nov 17. Virulence. 2015. PMID: 25483862 Free PMC article. Review.
Cited by
- Transcriptome analysis of bacteriophage communities in periodontal health and disease.
Santiago-Rodriguez TM, Naidu M, Abeles SR, Boehm TK, Ly M, Pride DT. Santiago-Rodriguez TM, et al. BMC Genomics. 2015 Jul 28;16(1):549. doi: 10.1186/s12864-015-1781-0. BMC Genomics. 2015. PMID: 26215258 Free PMC article. - Evaluation of bias induced by viral enrichment and random amplification protocols in metagenomic surveys of saliva DNA viruses.
Parras-Moltó M, Rodríguez-Galet A, Suárez-Rodríguez P, López-Bueno A. Parras-Moltó M, et al. Microbiome. 2018 Jun 28;6(1):119. doi: 10.1186/s40168-018-0507-3. Microbiome. 2018. PMID: 29954453 Free PMC article. - Sequence analysis of the human virome in febrile and afebrile children.
Wylie KM, Mihindukulasuriya KA, Sodergren E, Weinstock GM, Storch GA. Wylie KM, et al. PLoS One. 2012;7(6):e27735. doi: 10.1371/journal.pone.0027735. Epub 2012 Jun 13. PLoS One. 2012. PMID: 22719819 Free PMC article. - Genome Mining and Comparative Analysis of Streptococcus intermedius Causing Brain Abscess in a Child.
Issa E, Salloum T, Panossian B, Ayoub D, Abboud E, Tokajian S. Issa E, et al. Pathogens. 2019 Feb 13;8(1):22. doi: 10.3390/pathogens8010022. Pathogens. 2019. PMID: 30781742 Free PMC article. - New technologies for studying the complexity of oral diseases.
Burbelo PD, Bayat A, Lebovitz EE, Iadarola MJ. Burbelo PD, et al. Oral Dis. 2012 Mar;18(2):121-6. doi: 10.1111/j.1601-0825.2011.01863.x. Epub 2011 Oct 24. Oral Dis. 2012. PMID: 22023238 Free PMC article. Review.
References
- Hull MW, Chow AW. Indigenous microflora and innate immunity of the head and neck. Infect Dis Clin North Am. 2007;21:265–282. - PubMed
- Hentges DJ. The anaerobic microflora of the human body. Clin Infect Dis. 1993;16(Suppl 4):S175–S180. - PubMed
- Jenkinson HF, Lamont RJ. Oral microbial communities in sickness and in health. Trends Microbiol. 2005;13:589–595. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical