MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs - PubMed (original) (raw)
MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs
Robert J A Frost et al. Proc Natl Acad Sci U S A. 2010.
Abstract
Piwi-interacting RNAs (piRNAs) comprise a broad class of small noncoding RNAs that function as an endogenous defense system against transposable elements. Here we show that the putative DExD-box helicase MOV10-like-1 (MOV10L1) is essential for silencing retrotransposons in the mouse male germline. Mov10l1 is specifically expressed in germ cells with increasing expression from gonocytes/type A spermatogonia to pachytene spermatocytes. Primary spermatocytes of Mov10l1(-/-) mice show activation of LTR and LINE-1 retrotransposons, followed by cell death, causing male infertility and a complete block of spermatogenesis at early prophase of meiosis I. Despite the early expression of Mov10l1, germline stem cell maintenance appears unaffected in Mov10l1(-/-) mice. MOV10L1 interacts with the Piwi proteins MILI and MIWI. MOV10L1 also interacts with heat shock 70-kDa protein 2 (HSPA2), a testis-enriched chaperone expressed in pachytene spermatocytes and also essential for male fertility. These studies reveal a crucial role of MOV10L1 in male fertility and piRNA-directed retrotransposon silencing in male germ cells and suggest that MOV10L1 functions as a key component of a safeguard mechanism for the genetic information in male germ cells of mammals.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
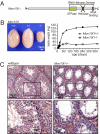
Mov10l1 is specifically expressed in spermatocytes within the testis. (A) Expression of Mov10l1, Mov10, and Mili mRNA in testes of C57BL/6 mice during postnatal development (GEO Series GES640). (B) Mov10l1 mRNA expression in fractions of undifferentiated (undiff.) spermatogonia and spermatocytes (diff.) compared with tubular and interstitial somatic testis cells isolated from testes of 19-d-old C57BL/6 mice (GEO series GES829). (C) Detection of Mov10l1 in embryonic (Upper) and juvenile/adult (Lower) testes by radioactive in situ hybridization with a probe directed against the C-terminal region of Mov10l1; exposure was 3 wk for embryonic and 1 week for postnatal tissue. (Upper Left) A dark-field image of a transverse section through the lower abdomen of an embryonic day 18 (E18) mouse. (Upper Center) An enlargement of Upper Left showing Mov10l1 mRNA as bright white dots in testis. (Upper Right) A high-magnification bright-field image of a seminiferous tubule depicting Mov10l1 as black silver grains localized specifically in gonocytes. (Lower) Detection of Mov10l1 in 7-wk-old adult (Lower Left, dark-field; Lower Center, bright-field) and in P15 juvenile (Lower Right) mouse testis. PS, pachytene spermatocytes; RS, round spermatids; ES, elongating spermatids; SG, spermatogonia.
Fig. 2.
Mov10l1−/− mice have a reduced testis size secondary to a lack of spermatids. (A) Scheme of the predicted MOV10L1 domains. For generation of Mov10l1−/− mice, the exon encoding the putative helicase domain (red) was flanked by LoxP sites, and global deletion of Mov10l1 was then achieved by breeding Mov10l1+/fl mice to CAG-Cre transgenic mice. (B) Mov10l1−/− mice showed reduced testis size compared with Mov10l1+/− and Mov10l1+/+ mice. (Left) Testes of 3-mo-old mice from the same litter. (Right) Testis weight of Mov10l1+/− and Mov10l1−/− mice. Each time point represents the mean of two to six testes. (C) Representative hematoxylin and eosin staining of transverse sections through testes of 3-mo-old wild-type versus Mov10l1−/− mice. (Lower) An enlargement of the regions indicated by the black frame in the Upper. ES, elongating spermatids; RS, round spermatids; PS, pachytene spermatocytes; DS, degenerating spermatocytes with condensed nuclei.
Fig. 3.
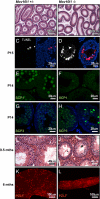
Infertility of Mov10l1−/− mice due to cell death of early pachytene spermatocytes. (A and B) Hematoxylin and eosin (H&E)-stained sections of epidydimides of 3-mo-old mice. Sperm were absent in epidydimides of Mov10l1−/− mice. (C and D) TUNEL staining of sections from testis of P15 Mov10l1+/− and Mov10l1−/− mice, respectively. (Left) Red channel with the apoptotic nuclei. (Right) Overlay with DAPI-stained nuclei (blue). (E–H) Confocal microscopy images of immunofluorescent staining of testis sections from P14-old mice for SCP1 (synaptonemal complex protein 1) and SCP3, respectively; counterstaining of nuclei with DAPI (blue) in G and H. (I and J) H&E staining of transverse sections of 9.5-mo-old mice; seminiferous tubules Mov10l1−/− mice still contained many spermatocytes and degenerating cells (arrowhead in J). (K and L) PZLF staining of tubules isolated from testes of 6-mo-old wild-type and Mov10l1−/− mice, respectively.
Fig. 4.
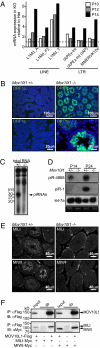
MOV10L1 interacts with Piwi proteins and is necessary for piRNA-dependent repression of retrotransposons. (A and B) Up-regulation of retrotransposons in Mov10l1−/− mice. (A) Microarray analysis of pooled total RNA (n = 3) from testes of Mov10l1−/− mice versus Mov10l1+/− mice at postnatal days 10, 12, and 14. The depicted retrotransposon families from the LINE and LTR class were among the strongest up-regulated transcripts in Mov10l1−/− mice at P14. (B) Immunofluorescent staining of testis sections from P14 mice for the LINE ORF1 protein (ORF1p, green); counterstaining of nuclei with DAPI (blue). (C) Total RNA of testis from 24-d-old Mov10l1+/− mice versus Mov10l1−/− mice was end-labeled with [32P]ATP and detected after denaturing PAGE. In Mov10l1−/− mice, ∼29-nucleotide-long RNAs, most likely representing piRNAs, were strongly reduced. (D) Total RNA from testis of P14 and P24 Mov10l1+/− versus Mov10l1−/− mice was isolated and used for Northern blotting to detect specific piRNAs (piR-4868 and piR-1) and miRNAs (let-7a). An ethidum bromide-stained polyacrylamide gel is shown as a loading control. (E) Detection of the Piwi proteins Mili and Miwi (white) in testis sections from P14 Mov10l1+/− versus Mov10l1−/− mice by immunofluorescent staining. (F) COS1 cells were cotransfected with expression plasmids for Flag-tagged MOV10L1 and Myc-tagged MILI and MIWI, respectively. Lysates were then used for coimmunoprecipitation with anti-Flag antibodies followed by Western blot analysis with anti-Myc antibodies. Input lanes represent 1% of lysates used for coimmunoprecipitation. The experiment was repeated three times; representative immunoblots are shown. Reciprocal coimmunoprecipitation worked similarly.
Fig. 5.
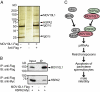
HSPA2 is an interaction partner of MOV10L1. (A) Identification of MOV10L1-interacting partners by pull-down experiment with recombinant MOV10L1. Forty-eight hours before harvesting, COS cells were Mock-transfected (left lane) or transfected with a plasmid expressing Flag-tagged MOV10L1 (center and right lanes). Lysates were then incubated with anti-Flag antibody and Sepharose A (left and right lanes) or only with Sepharose A (center lane). Flag-tagged MOV10L1 bound to the Sepharose was then incubated with lysates from P20 mouse testis. Proteins bound to MOV10L1 and Sepharose were separated by SDS-PAGE. After silver staining of the gel, protein bands that were visible only in the right lane, but not in the two control lanes, were cut out and analyzed by MALDI-TOF mass spectrometry. HSPA2, heat shock 70-kDa protein 2; IgG, Ig; hc, heavy chain; lc, light chain. (B) COS1 cells were cotransfected with expression plasmids for Flag-tagged MOV10L1 and Myc-tagged HSPA2. Lysates were then used for coimmunoprecipitation with anti-Flag antibodies followed by Western blot analysis with anti-Myc antibodies. Input lanes represent 1% of lysates used for coimmunoprecipitation. Reciprocal coimmunoprecipitation worked similarily. (C) Model for the role of MOV10L1 during spermatogenesis. MOV10L1 interacts with MILI and MIWI, which is necessary for piRNA-induced repression of retrotransposons in pachytene spermatocytes. MOV10L1 interacts further with HSPA2.
Similar articles
- Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway.
Zheng K, Xiol J, Reuter M, Eckardt S, Leu NA, McLaughlin KJ, Stark A, Sachidanandam R, Pillai RS, Wang PJ. Zheng K, et al. Proc Natl Acad Sci U S A. 2010 Jun 29;107(26):11841-6. doi: 10.1073/pnas.1003953107. Epub 2010 Jun 1. Proc Natl Acad Sci U S A. 2010. PMID: 20534472 Free PMC article. - yama, a mutant allele of Mov10l1, disrupts retrotransposon silencing and piRNA biogenesis.
Guan Y, Keeney S, Jain D, Wang PJ. Guan Y, et al. PLoS Genet. 2021 Feb 26;17(2):e1009265. doi: 10.1371/journal.pgen.1009265. eCollection 2021 Feb. PLoS Genet. 2021. PMID: 33635934 Free PMC article. - Blockade of pachytene piRNA biogenesis reveals a novel requirement for maintaining post-meiotic germline genome integrity.
Zheng K, Wang PJ. Zheng K, et al. PLoS Genet. 2012;8(11):e1003038. doi: 10.1371/journal.pgen.1003038. Epub 2012 Nov 15. PLoS Genet. 2012. PMID: 23166510 Free PMC article. - MOV10L1 in piRNA processing and gene silencing of retrotransposons during spermatogenesis.
Zhu X, Zhi E, Li Z. Zhu X, et al. Reproduction. 2015 May;149(5):R229-35. doi: 10.1530/REP-14-0569. Epub 2015 Feb 9. Reproduction. 2015. PMID: 25667429 Review. - piRNA and spermatogenesis in mice.
Chuma S, Nakano T. Chuma S, et al. Philos Trans R Soc Lond B Biol Sci. 2013 Jan 5;368(1609):20110338. doi: 10.1098/rstb.2011.0338. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23166399 Free PMC article. Review.
Cited by
- Impact of heat shock transcription factor 1 on global gene expression profiles in cells which induce either cytoprotective or pro-apoptotic response following hyperthermia.
Kus-Liśkiewicz M, Polańska J, Korfanty J, Olbryt M, Vydra N, Toma A, Widłak W. Kus-Liśkiewicz M, et al. BMC Genomics. 2013 Jul 8;14:456. doi: 10.1186/1471-2164-14-456. BMC Genomics. 2013. PMID: 23834426 Free PMC article. - Mouse EWSR1 is crucial for spermatid post-meiotic transcription and spermiogenesis.
Tian H, Petkov PM. Tian H, et al. Development. 2021 Jun 1;148(11):dev199414. doi: 10.1242/dev.199414. Epub 2021 Jun 8. Development. 2021. PMID: 34100066 Free PMC article. - DNA methylation signature in peripheral blood reveals distinct characteristics of human X chromosome numerical aberrations.
Sharma A, Jamil MA, Nuesgen N, Schreiner F, Priebe L, Hoffmann P, Herns S, Nöthen MM, Fröhlich H, Oldenburg J, Woelfle J, El-Maarri O. Sharma A, et al. Clin Epigenetics. 2015 Jul 28;7(1):76. doi: 10.1186/s13148-015-0112-2. eCollection 2015. Clin Epigenetics. 2015. PMID: 26221191 Free PMC article. - Incomplete cre-mediated excision leads to phenotypic differences between Stra8-iCre; Mov10l1(lox/lox) and Stra8-iCre; Mov10l1(lox/Δ) mice.
Bao J, Ma HY, Schuster A, Lin YM, Yan W. Bao J, et al. Genesis. 2013 Jul;51(7):481-90. doi: 10.1002/dvg.22389. Epub 2013 Mar 30. Genesis. 2013. PMID: 23554062 Free PMC article. - Coupled protein synthesis and ribosome-guided piRNA processing on mRNAs.
Sun YH, Wang RH, Du K, Zhu J, Zheng J, Xie LH, Pereira AA, Zhang C, Ricci EP, Li XZ. Sun YH, et al. Nat Commun. 2021 Oct 13;12(1):5970. doi: 10.1038/s41467-021-26233-8. Nat Commun. 2021. PMID: 34645830 Free PMC article.
References
- Babushok DV, Kazazian HH., Jr Progress in understanding the biology of the human mutagen LINE-1. Hum Mutat. 2007;28:527–539. - PubMed
- Kazazian HH., Jr Mobile elements: Drivers of genome evolution. Science. 2004;303:1626–1632. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HD036022-10/HD/NICHD NIH HHS/United States
- R01 HD036022/HD/NICHD NIH HHS/United States
- R01 HD053889/HD/NICHD NIH HHS/United States
- R01 HL093039/HL/NHLBI NIH HHS/United States
- R01 HD036022-09/HD/NICHD NIH HHS/United States
- R01 HD061575/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials