GPU-enabled FREALIGN: accelerating single particle 3D reconstruction and refinement in Fourier space on graphics processors - PubMed (original) (raw)
GPU-enabled FREALIGN: accelerating single particle 3D reconstruction and refinement in Fourier space on graphics processors
Xueming Li et al. J Struct Biol. 2010 Dec.
Abstract
Among all the factors that determine the resolution of a 3D reconstruction by single particle electron cryo-microscopy (cryoEM), the number of particle images used in the dataset plays a major role. More images generally yield better resolution, assuming the imaged protein complex is conformationally and compositionally homogeneous. To facilitate processing of very large datasets, we modified the computer program, FREALIGN, to execute the computationally most intensive procedures on Graphics Processing Units (GPUs). Using the modified program, the execution speed increased between 10 and 240-fold depending on the task performed by FREALIGN. Here we report the steps necessary to parallelize critical FREALIGN subroutines and evaluate its performance on computers with multiple GPUs.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures
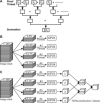
Figure 1. Implementing parallelization of FREALIGN
(A) Tree-structured parallel summation algorithm. The sums between every two pixels within the same block are computed in synchronization, and repeated until the final result is obtained. (B) Dividing and distributing subsets of the data on multiple GPUs for search and refinement. (C) Dividing and distributing subsets of the data on multiple GPUs for 3D reconstruction.
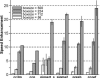
Figure 2. Speed enhancement factors of individual FREALIGN subroutines
Acceleration of individual subroutines with different image sizes: 56 × 56 to 500 × 500 pixels. For the measurement of the speed enhancement factors, only one GPU on system I was used.
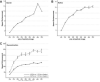
Figure 3. Acceleration of search and refinement by GPU-enabled FREALIGN with various image sizes
(A) Search and (B) refinement. The accelerations factors were measured on a single GPU on System I vs. a single i7 CPU core on the same system. The corresponding execution times are listed in Supplementary Table 3 and 4. (C) Reconstruction. The acceleration factors were measured on 4 and 8 GPUs on System I and II, respectively, vs. a single CPU core on the same systems.
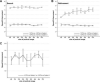
Figure 4. Acceleration of GPU-enabled FREALIGN on multiple GPUs
(A) Search, (B) refinement and (C) 3D reconstruction. Solid lines are the speed enhancement factors obtained for System I while dashed lines apply to System II. All corresponding data are listed in Supplementary Table 5.
Similar articles
- Gctf: Real-time CTF determination and correction.
Zhang K. Zhang K. J Struct Biol. 2016 Jan;193(1):1-12. doi: 10.1016/j.jsb.2015.11.003. Epub 2015 Nov 19. J Struct Biol. 2016. PMID: 26592709 Free PMC article. - Frealign: An Exploratory Tool for Single-Particle Cryo-EM.
Grigorieff N. Grigorieff N. Methods Enzymol. 2016;579:191-226. doi: 10.1016/bs.mie.2016.04.013. Epub 2016 Jun 7. Methods Enzymol. 2016. PMID: 27572728 Free PMC article. Review. - Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2.
Kimanius D, Forsberg BO, Scheres SH, Lindahl E. Kimanius D, et al. Elife. 2016 Nov 15;5:e18722. doi: 10.7554/eLife.18722. Elife. 2016. PMID: 27845625 Free PMC article. - Symmetry-adapted spherical harmonics method for high-resolution 3D single-particle reconstructions.
Liu H, Cheng L, Zeng S, Cai C, Zhou ZH, Yang Q. Liu H, et al. J Struct Biol. 2008 Jan;161(1):64-73. doi: 10.1016/j.jsb.2007.09.016. Epub 2007 Oct 1. J Struct Biol. 2008. PMID: 17977017 - Cryo-EM Structure Determination Using Segmented Helical Image Reconstruction.
Fromm SA, Sachse C. Fromm SA, et al. Methods Enzymol. 2016;579:307-28. doi: 10.1016/bs.mie.2016.05.034. Epub 2016 Jun 28. Methods Enzymol. 2016. PMID: 27572732 Review.
Cited by
- Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM.
Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. Li X, et al. Nat Methods. 2013 Jun;10(6):584-90. doi: 10.1038/nmeth.2472. Epub 2013 May 5. Nat Methods. 2013. PMID: 23644547 Free PMC article. - Advances in methods for atomic resolution macromolecular structure determination.
Thompson MC, Yeates TO, Rodriguez JA. Thompson MC, et al. F1000Res. 2020 Jul 2;9:F1000 Faculty Rev-667. doi: 10.12688/f1000research.25097.1. eCollection 2020. F1000Res. 2020. PMID: 32676184 Free PMC article. Review. - Asynchronous data acquisition and on-the-fly analysis of dose fractionated cryoEM images by UCSFImage.
Li X, Zheng S, Agard DA, Cheng Y. Li X, et al. J Struct Biol. 2015 Nov;192(2):174-8. doi: 10.1016/j.jsb.2015.09.003. Epub 2015 Sep 11. J Struct Biol. 2015. PMID: 26370395 Free PMC article. - Nectin-like interactions between poliovirus and its receptor trigger conformational changes associated with cell entry.
Strauss M, Filman DJ, Belnap DM, Cheng N, Noel RT, Hogle JM. Strauss M, et al. J Virol. 2015 Apr;89(8):4143-57. doi: 10.1128/JVI.03101-14. Epub 2015 Jan 28. J Virol. 2015. PMID: 25631086 Free PMC article. - gEMpicker: a highly parallel GPU-accelerated particle picking tool for cryo-electron microscopy.
Hoang TV, Cavin X, Schultz P, Ritchie DW. Hoang TV, et al. BMC Struct Biol. 2013 Oct 21;13:25. doi: 10.1186/1472-6807-13-25. BMC Struct Biol. 2013. PMID: 24144335 Free PMC article.
References
- Bilbao-Castro JR, Marabini R, Sorzano CO, Garcia I, Carazo JM, Fernandez JJ. Exploiting desktop supercomputing for three-dimensional electron microscopy reconstructions using ART with blobs. J Struct Biol. 2009;165:19–26. - PubMed
- Castano Diez D, Mueller H, Frangakis AS. Implementation and performance evaluation of reconstruction algorithms on graphics processors. J Struct Biol. 2007;157:288–95. - PubMed
- Castano-Diez D, Moser D, Schoenegger A, Pruggnaller S, Frangakis AS. Performance evaluation of image processing algorithms on the GPU. J Struct Biol. 2008;164:153–60. - PubMed
- Cheng Y, Zak O, Aisen P, Harrison SC, Walz T. Structure of the human transferrin receptor-transferrin complex. Cell. 2004;116:565–76. - PubMed
Publication types
MeSH terms
Grants and funding
- R01GM082893/GM/NIGMS NIH HHS/United States
- P01 GM62580/GM/NIGMS NIH HHS/United States
- 1S10RR026814-01/RR/NCRR NIH HHS/United States
- P50GM082250/GM/NIGMS NIH HHS/United States
- S10 RR026814-01/RR/NCRR NIH HHS/United States
- P50 GM082250/GM/NIGMS NIH HHS/United States
- P01 GM062580-050002/GM/NIGMS NIH HHS/United States
- S10 RR026814/RR/NCRR NIH HHS/United States
- R01 GM082893/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 GM082893-02/GM/NIGMS NIH HHS/United States
- P01 GM062580/GM/NIGMS NIH HHS/United States
- P50 GM082250-03/GM/NIGMS NIH HHS/United States
- P50 GM082250-039001/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources