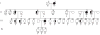Clinical, neuroimaging and neuropathological features of a new chromosome 9p-linked FTD-ALS family - PubMed (original) (raw)
. 2011 Feb;82(2):196-203.
doi: 10.1136/jnnp.2009.204081. Epub 2010 Jun 20.
Ian R Mackenzie, Bradley F Boeve, Matthew Baker, William W Seeley, Richard Crook, Howard Feldman, Ging-Yuek R Hsiung, Nicola Rutherford, Victor Laluz, Jennifer Whitwell, Dean Foti, Eric McDade, Jennifer Molano, Anna Karydas, Aleksandra Wojtas, Jill Goldman, Jacob Mirsky, Pheth Sengdy, Stephen Dearmond, Bruce L Miller, Rosa Rademakers
Affiliations
- PMID: 20562461
- PMCID: PMC3017222
- DOI: 10.1136/jnnp.2009.204081
Clinical, neuroimaging and neuropathological features of a new chromosome 9p-linked FTD-ALS family
Adam L Boxer et al. J Neurol Neurosurg Psychiatry. 2011 Feb.
Abstract
Background: Frontotemporal dementia-amyotrophic lateral sclerosis (FTD-ALS) is a heritable form of FTD, but the gene(s) responsible for the majority of autosomal dominant FTD-ALS cases have yet to be found. Previous studies have identified a region on chromosome 9p that is associated with FTD and ALS.
Methods: The authors report the clinical, volumetric MRI, neuropathological and genetic features of a new chromosome 9p-linked FTD-ALS family, VSM-20.
Results: Ten members of family VSM-20 displayed heterogeneous clinical phenotypes of isolated behavioural-variant FTD (bvFTD), ALS or a combination of the two. Parkinsonism was common, with one individual presenting with a corticobasal syndrome. Analysis of structural MRI scans from five affected family members revealed grey- and white-matter loss that was most prominent in the frontal lobes, with mild parietal and occipital lobe atrophy, but less temporal lobe atrophy than in 10 severity-matched sporadic bvFTD cases. Autopsy in three family members showed a consistent and unique subtype of FTLD-TDP pathology. Genome-wide linkage analysis conclusively linked family VSM-20 to a 28.3 cM region between D9S1808 and D9S251 on chromosome 9p, reducing the published minimal linked region to a 3.7 Mb interval. Genomic sequencing and expression analysis failed to identify mutations in the 10 known and predicted genes within this candidate region, suggesting that next-generation sequencing may be needed to determine the mutational mechanism associated with chromosome 9p-linked FTD-ALS.
Conclusions: Family VSM-20 significantly reduces the region linked to FTD-ALS on chromosome 9p. A distinct pattern of brain atrophy and neuropathological findings may help to identify other families with FTD-ALS caused by this genetic abnormality.
Conflict of interest statement
Competing interests None.
Figures
Figure 1
Pedigree of family VSM-20. Black symbols represent patients affected with behavioural-variant frontotemporal dementia (left side filled), amyotrophic lateral sclerosis (right side filled) or both. White symbols represent unaffected individuals or at-risk individuals with unknown phenotype. The Roman numeral to the left of the pedigree denotes the generation; Arabic numbers above the individuals denote individuals. The Arabic numbers below the individuals denote age at onset for patients and either age at last examination or age at death for unaffected or at-risk individuals with unknown phenotype. The ‘+’ sign indicates individuals included in the linkage analysis. For generation IV, only individuals included in the linkage analysis are shown.
Figure 2
Patterns of brain atrophy in two VSM-20 clinical phenotypes: coronal T1-weighted MRI section at MNI coordinate y=14, and Freesurfer-derived cortical thickness maps in (A) a 54-year-old behavioural-variant frontotemporal dementia subject (II-8) as compared with 20 male normal control subjects (mean age 55.5) and (B) a 40-year-old pure amyotrophic lateral sclerosis subject (III-9) as compared with 25 female normal control subjects (mean age 59.8) displayed on rendered normal control MRI template. Coloured areas indicate cortical thickness reductions p<0.05 to p<0.001, uncorrected, versus controls.
Figure 3
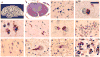
Post-mortem neuropathology in affected members of VMS-20. Cerebral atrophy was most consistent in the frontal lobes (A, case II-8). The corticospinal tracts showed loss of myelin staining (B, case III-7). Ubiquitin- and TDP-43-immunoreactive neuronal cytoplasmic inclusions (NCI) were present in all layers of the frontal and temporal neocortex (arrows, C) and either had a smooth round contour or appeared to be an aggregate of coarse granules (D). TDP-43-positive granular preinclusions were most common in the upper neocortical layers (E). Swollen dystrophic neurites in the neocortex were best demonstrated with ubiquitin immunoistochemistry (F). Moderate numbers of TDP-43-immunoreactive glial cytoplasmic inclusions were present in the subcortical white matter (G). In the hippocampus, compact neuronal cytoplasmic inclusions (NCI) were numerous in the dentate granule cells (H), while hippocampal pyramidal neurons often contained more ill-defined NCI (I). NCI were present in lower motor neurons of the brainstem and spinal cord, and had a granular, filamentous (J) or compact (K) morphology. There were numerous round NCI and neurites in the cerebellar granular layer that were immunoreactive ubiquitin but not TDP-43 (L). (A) gross photograph; (B) haematoxylin and eosin with luxol fast blue; (C, F, H–L), ubiquitin immunohistochemistry; (D, E and G) TDP-43 immunohistochemistry. Scale bar: (A) 6 cm; (B) 300 μm; (C, F, H and I) 50 μm; (D, E and G) 12 μm; (J and K) 30 μm; (L) 20 μm.
Figure 4
Disguised linkage pedigree of family VSM-20. Black symbols represent patients affected with behavioural-variant frontotemporal dementia (left side filled), amyotrophic lateral sclerosis (right side filled) or both. White symbols represent unaffected individuals or at-risk individuals with unknown phenotype. The Roman numeral to the left of the pedigree denotes the generation; Arabic numbers above the individuals denote individuals. Haplotypes are based on 16 informative markers at chromosome 9p. Haplotypes for individuals I-1, I-2, II-3 and II-4 are inferred from genotype data of siblings and offspring. Detailed pedigree is shown in Figure 1.
Figure 5
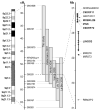
Schematic presentation of chromosome 9p FTD-ALS locus. Grey bars indicate the minimal candidate regions on chromosome 9p in family VSM-20 and all other previously reported 9p-linked FTD-ALS families. Together, these studies define a 3.7 Mb interval between D9S169 and D9S251 containing 10 genes (shown in detail on the right). The five protein coding genes analysed in detail in this study are highlighted in bold.
Similar articles
- Familial frontotemporal dementia with amyotrophic lateral sclerosis and a shared haplotype on chromosome 9p.
Pearson JP, Williams NM, Majounie E, Waite A, Stott J, Newsway V, Murray A, Hernandez D, Guerreiro R, Singleton AB, Neal J, Morris HR. Pearson JP, et al. J Neurol. 2011 Apr;258(4):647-55. doi: 10.1007/s00415-010-5815-x. Epub 2010 Nov 12. J Neurol. 2011. PMID: 21072532 Free PMC article. - Clinical and pathological features of familial frontotemporal dementia caused by C9ORF72 mutation on chromosome 9p.
Hsiung GY, DeJesus-Hernandez M, Feldman HH, Sengdy P, Bouchard-Kerr P, Dwosh E, Butler R, Leung B, Fok A, Rutherford NJ, Baker M, Rademakers R, Mackenzie IR. Hsiung GY, et al. Brain. 2012 Mar;135(Pt 3):709-22. doi: 10.1093/brain/awr354. Epub 2012 Feb 17. Brain. 2012. PMID: 22344582 Free PMC article. - Frontotemporal dementia-amyotrophic lateral sclerosis syndrome locus on chromosome 16p12.1-q12.2: genetic, clinical and neuropathological analysis.
Dobson-Stone C, Luty AA, Thompson EM, Blumbergs P, Brooks WS, Short CL, Field CD, Panegyres PK, Hecker J, Solski JA, Blair IP, Fullerton JM, Halliday GM, Schofield PR, Kwok JB. Dobson-Stone C, et al. Acta Neuropathol. 2013 Apr;125(4):523-33. doi: 10.1007/s00401-013-1078-9. Epub 2013 Jan 22. Acta Neuropathol. 2013. PMID: 23338750 Free PMC article. - The neuropathology of FTD associated With ALS.
Mackenzie IR. Mackenzie IR. Alzheimer Dis Assoc Disord. 2007 Oct-Dec;21(4):S44-9. doi: 10.1097/WAD.0b013e31815c3486. Alzheimer Dis Assoc Disord. 2007. PMID: 18090423 Review. - Making connections: pathology and genetics link amyotrophic lateral sclerosis with frontotemporal lobe dementia.
Fecto F, Siddique T. Fecto F, et al. J Mol Neurosci. 2011 Nov;45(3):663-75. doi: 10.1007/s12031-011-9637-9. Epub 2011 Sep 7. J Mol Neurosci. 2011. PMID: 21901496 Review.
Cited by
- Characterization of frontotemporal dementia and/or amyotrophic lateral sclerosis associated with the GGGGCC repeat expansion in C9ORF72.
Boeve BF, Boylan KB, Graff-Radford NR, DeJesus-Hernandez M, Knopman DS, Pedraza O, Vemuri P, Jones D, Lowe V, Murray ME, Dickson DW, Josephs KA, Rush BK, Machulda MM, Fields JA, Ferman TJ, Baker M, Rutherford NJ, Adamson J, Wszolek ZK, Adeli A, Savica R, Boot B, Kuntz KM, Gavrilova R, Reeves A, Whitwell J, Kantarci K, Jack CR Jr, Parisi JE, Lucas JA, Petersen RC, Rademakers R. Boeve BF, et al. Brain. 2012 Mar;135(Pt 3):765-83. doi: 10.1093/brain/aws004. Brain. 2012. PMID: 22366793 Free PMC article. - Neuropsychiatric features of C9orf72-associated behavioral variant frontotemporal dementia and frontotemporal dementia with motor neuron disease.
Takada LT, Sha SJ. Takada LT, et al. Alzheimers Res Ther. 2012 Oct 3;4(5):38. doi: 10.1186/alzrt141. eCollection 2012. Alzheimers Res Ther. 2012. PMID: 23034079 Free PMC article. Review. - RNA-mediated toxicity in neurodegenerative disease.
Belzil VV, Gendron TF, Petrucelli L. Belzil VV, et al. Mol Cell Neurosci. 2013 Sep;56:406-19. doi: 10.1016/j.mcn.2012.12.006. Epub 2012 Dec 29. Mol Cell Neurosci. 2013. PMID: 23280309 Free PMC article. Review. - Frontotemporal dementia with psychosis, parkinsonism, visuo-spatial dysfunction, upper motor neuron involvement associated to expansion of C9ORF72: a peculiar phenotype?
Floris G, Borghero G, Cannas A, Di Stefano F, Costantino E, Murru MR, Brunetti M, Restagno G, Traynor BJ, Marrosu MG, Chiò A, Marrosu F. Floris G, et al. J Neurol. 2012 Aug;259(8):1749-51. doi: 10.1007/s00415-012-6444-3. Epub 2012 Feb 10. J Neurol. 2012. PMID: 22323211 Free PMC article. No abstract available. - C9ORF72 Gene GGGGCC Hexanucleotide Expansion: A High Clinical Variability from Amyotrophic Lateral Sclerosis to Frontotemporal Dementia.
Kortazar-Zubizarreta I, Manero-Azua A, Afonso-Agüera J, Perez de Nanclares G. Kortazar-Zubizarreta I, et al. J Pers Med. 2023 Sep 19;13(9):1396. doi: 10.3390/jpm13091396. J Pers Med. 2023. PMID: 37763163 Free PMC article.
References
- Lomen-Hoerth C, Anderson T, Miller B. The overlap of amyotrophic lateral sclerosis and frontotemporal dementia. Neurology. 2002;59:1077–9. - PubMed
- Hodges JR, Davies R, Xuereb J, et al. Survival in frontotemporal dementia. Neurology. 2003;61:349–54. - PubMed
- Roberson ED, Hesse JH, Rose KD, et al. Frontotemporal dementia progresses to death faster than Alzheimer disease. Neurology. 2005;65:719–25. - PubMed
- Chang JL, Lomen-Hoerth C, Murphy J, et al. A voxel-based morphometry study of patterns of brain atrophy in ALS and ALS/FTLD. Neurology. 2005;65:75–80. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 AG031278/AG/NIA NIH HHS/United States
- P50 AG016574/AG/NIA NIH HHS/United States
- K23 NS048855/NS/NINDS NIH HHS/United States
- R01 AG031278-02/AG/NIA NIH HHS/United States
- P01 AG019724/AG/NIA NIH HHS/United States
- R01 NS065782/NS/NINDS NIH HHS/United States
- P01AG019724/AG/NIA NIH HHS/United States
- K23NS48855/NS/NINDS NIH HHS/United States
- CAPMC/ CIHR/Canada
- R01AG031278/AG/NIA NIH HHS/United States
- K23 NS048855-05/NS/NINDS NIH HHS/United States
- P50AG16574/AG/NIA NIH HHS/United States
- P50AG0300601/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous