RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation - PubMed (original) (raw)
RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation
Iván D'Orso et al. Nat Struct Mol Biol. 2010 Jul.
Abstract
The transition from transcription initiation to elongation at the HIV-1 promoter is controlled by Tat, which recruits P-TEFb to TAR RNA to phosphorylate RNA polymerase II. It has long been unclear why the HIV-1 promoter is incompetent for elongation. We report that P-TEFb is recruited to the promoter in a catalytically inactive state bound to the inhibitory 7SK small nuclear ribonucleoprotein (snRNP), thereby preventing elongation. It also has long been believed that TAR functions to recruit Tat to the promoter, but we find that Tat is recruited to the DNA template before TAR is synthesized. We propose that TAR binds Tat and P-TEFb as it emerges on the nascent transcript, competitively displacing the inhibitory 7SK snRNP and activating the P-TEFb kinase. Recruitment of an inhibitory snRNP complex at an early stage in the transcription cycle provides a new paradigm for controlling gene expression with a noncoding RNA.
Figures
Figure 1
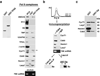
Inactive P-TEFb assembles into Pol II complexes and Tat activates its catalytic activity. (a) Partial composition of Pol II complexes purified on a TFIIS affinity resin was assessed by comparing Western blots of proteins and levels of 7SK snRNA eluted from a GST-TFIIS column to those from HeLa whole cell extracts (WCE) and a GST control column. The TFIIS used was an N-terminal fragment that possesses the same activity as full-length TFIIS. The purified coomassie-stained recombinant GST and GST-TFIIS proteins used for the columns are shown on the left. The arrow indicates the position of full-length CycT1. (b) GST-TFIIS eluted complexes were loaded onto a Sepharose CL-2B gel filtration column and the single eluted peak was immunoprecipitated (IP) by either anti-Cdk9 or mock (normal rabbit IgG) antibodies. Both precipitates, either incubated or not with purified GST-Tat, were used in kinase assays using a GST-CTD substrate and analyzed by Western blot with the H5 antibody, which recognizes Pol IIo. (c) Pol II complexes were purified on a TFIIS affinity resin, incubated with GST or GST-Tat in vitro, immunoprecipitated with anti-Cdk9 antibody, and the presence of P-TEFb (CycT1 and Cdk9) and 7SK snRNP (Hexim1 and Larp7) was monitored by Western blot.
Figure 2
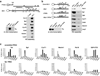
Distribution of Tat and cofactors at the HIV-1 promoter and dependence on TAR. (a) Schematic of the HIV-1 LTR promoter-luciferase (FFL) reporter integrated into a HeLa cell line, showing the locations of the upstream region (−845), enhancer elements (−352), core promoter containing Sp1 and TATA-boxes (−75), transcription start site (TSS; +1), TAR element, FFL coding region, stop codon, and polyadenylation signal (p(A)). The locations of eight amplicons used in PCR quantification of ChIP-enriched DNA are shown. Numbers indicate the positions of the central base pair of each amplicon relative to the TSS. (b) ChIP assays were performed with protein extracts from the reporter cell line 48 hr after a mock (grey bars) or Flag-tagged Tat transfection (black bars) using the antibodies indicated. Values represent the percentage of input DNA immunoprecipitated and are the average of four independent PCRs from two separate immunoprecipitations from two independent cell cultures. All standard deviations are <15%. (c) ChIP assays were performed as in panel b but using extracts pre-treated with RNase A. (d) ChIP assays were performed with protein extracts obtained from a HeLa HIV-1 LTRΔTAR-FFL cell line 48 hr after a mock (grey bars) or Flag-tagged Tat transfection (black bars) using the antibodies indicated.
Figure 3
Tat assembles with the 7SK snRNP in vivo. RNA immunoprecipitation (RIP) was performed using antibodies to Flag-tagged Tat. Western blots (upper panels) indicate that components of P-TEFb (CycT1 and Cdk9) and 7SK snRNP (Larp7 and Hexim1) form a complex with Tat. The two lower panels utilized RT-PCR to detect 7SK and U6 snRNAs.
Figure 4
7SK snRNP and Tat recruitment to HIV-1 PICs are Sp1-dependent. (a) Promoterless and full-length LTR templates were immobilized to streptavidin-coated magnetic beads through a biotin moiety at the 5’ end, and a NotI site was used to cleave the promoter and elute proteins from the beads, which were then analyzed by Western blot. The activity of the purified HIV-1 PICs was measured by in vitro transcription and primer extension and yielded the expected transcription product (Tp), and was inhibited by α-amanitin treatment. (b) The immobilized full-length HIV-1 LTR, core, and mutant templates shown were incubated with HeLa nuclear extracts and washed, and proteins were eluted with NotI and identified by Western blot. In vitro transcription of the PICs produced the correct Tp. (c) HeLa HIV-1 LTR reporter cells were transfected with a control scrambled siRNA or a siRNA against Sp1, resulting in >80% depletion of Sp1 and decrease in reporter activity (Supplementary Fig. 5), and ChIP assays were performed using the antibodies indicated. Values represent the percentage of input DNA immunoprecipitated and are the average of four independent PCRs from two separate immunoprecipitations from two independent cell cultures. All standard deviations are <15%
Figure 5
Proposed model of HIV-1 transcription activation by Tat. (a) Tat assembles into complexes with P-TEFb (CycT1 and Cdk9) and the 7SK snRNP (Hexim1, Larp7, and 7SK snRNA). This Tat-7SK snRNP complex is recruited to HIV-1 PICs containing the Pol IIa form and the basal transcription machinery (e.g. Sp1, TBP), among other possible promoter-specific factors, and remains bound in the paused state (Paused complex). As transcription proceeds and TAR is synthesized, Tat facilitates the transfer of P-TEFb to the nascent RNA site. We propose that this Tat-TAR binding event competitively displaces 7SK snRNP and activates Cdk9 to phosphorylate Ser2 residues in the CTD (P) and assemble competent transcription elongation complexes containing a Pol IIo form. Hexim1 may dissociate from Larp7/7SK snRNA complexes, as it is not stably bound, and may be replaced by hnRNP proteins in a transcription-dependent manner,. (b) In the absence of TAR, Tat and P-TEFb do not transfer to the nascent RNA and evict the 7SK snRNP, preventing Ser2-CTD phosphorylation (P) and formation of elongation complexes.
Comment in
- Kick-sTARting HIV-1 transcription elongation by 7SK snRNP deporTATion.
Barboric M, Lenasi T. Barboric M, et al. Nat Struct Mol Biol. 2010 Aug;17(8):928-30. doi: 10.1038/nsmb0810-928. Nat Struct Mol Biol. 2010. PMID: 20683478
Similar articles
- Transition step during assembly of HIV Tat:P-TEFb transcription complexes and transfer to TAR RNA.
D'Orso I, Jang GM, Pastuszak AW, Faust TB, Quezada E, Booth DS, Frankel AD. D'Orso I, et al. Mol Cell Biol. 2012 Dec;32(23):4780-93. doi: 10.1128/MCB.00206-12. Epub 2012 Sep 24. Mol Cell Biol. 2012. PMID: 23007159 Free PMC article. - Controlling cellular P-TEFb activity by the HIV-1 transcriptional transactivator Tat.
Muniz L, Egloff S, Ughy B, Jády BE, Kiss T. Muniz L, et al. PLoS Pathog. 2010 Oct 14;6(10):e1001152. doi: 10.1371/journal.ppat.1001152. PLoS Pathog. 2010. PMID: 20976203 Free PMC article. - The HIV-1 Tat protein recruits a ubiquitin ligase to reorganize the 7SK snRNP for transcriptional activation.
Faust TB, Li Y, Bacon CW, Jang GM, Weiss A, Jayaraman B, Newton BW, Krogan NJ, D'Orso I, Frankel AD. Faust TB, et al. Elife. 2018 May 30;7:e31879. doi: 10.7554/eLife.31879. Elife. 2018. PMID: 29845934 Free PMC article. - New insights into the control of HIV-1 transcription: when Tat meets the 7SK snRNP and super elongation complex (SEC).
He N, Zhou Q. He N, et al. J Neuroimmune Pharmacol. 2011 Jun;6(2):260-8. doi: 10.1007/s11481-011-9267-6. Epub 2011 Mar 1. J Neuroimmune Pharmacol. 2011. PMID: 21360054 Free PMC article. Review. - Cracking the control of RNA polymerase II elongation by 7SK snRNP and P-TEFb.
C Quaresma AJ, Bugai A, Barboric M. C Quaresma AJ, et al. Nucleic Acids Res. 2016 Sep 19;44(16):7527-39. doi: 10.1093/nar/gkw585. Epub 2016 Jul 1. Nucleic Acids Res. 2016. PMID: 27369380 Free PMC article. Review.
Cited by
- Protein intrinsic disorder as a flexible armor and a weapon of HIV-1.
Xue B, Mizianty MJ, Kurgan L, Uversky VN. Xue B, et al. Cell Mol Life Sci. 2012 Apr;69(8):1211-59. doi: 10.1007/s00018-011-0859-3. Epub 2011 Oct 28. Cell Mol Life Sci. 2012. PMID: 22033837 Free PMC article. Review. - Nascent RNA: Friend or foe of the chromatin bound?
Hyder U, D'Orso I. Hyder U, et al. Mol Cell. 2021 Jul 15;81(14):2871-2872. doi: 10.1016/j.molcel.2021.06.018. Mol Cell. 2021. PMID: 34270942 Free PMC article. - Stochastic pausing at latent HIV-1 promoters generates transcriptional bursting.
Tantale K, Garcia-Oliver E, Robert MC, L'Hostis A, Yang Y, Tsanov N, Topno R, Gostan T, Kozulic-Pirher A, Basu-Shrivastava M, Mukherjee K, Slaninova V, Andrau JC, Mueller F, Basyuk E, Radulescu O, Bertrand E. Tantale K, et al. Nat Commun. 2021 Jul 23;12(1):4503. doi: 10.1038/s41467-021-24462-5. Nat Commun. 2021. PMID: 34301927 Free PMC article. - Functional Segregation of Overlapping Genes in HIV.
Fernandes JD, Faust TB, Strauli NB, Smith C, Crosby DC, Nakamura RL, Hernandez RD, Frankel AD. Fernandes JD, et al. Cell. 2016 Dec 15;167(7):1762-1773.e12. doi: 10.1016/j.cell.2016.11.031. Cell. 2016. PMID: 27984726 Free PMC article. - The ARF tumor suppressor targets PPM1G/PP2Cγ to counteract NF-κB transcription tuning cell survival and the inflammatory response.
Hyder U, McCann JL, Wang J, Fung V, Bayo J, D'Orso I. Hyder U, et al. Proc Natl Acad Sci U S A. 2020 Dec 22;117(51):32594-32605. doi: 10.1073/pnas.2004470117. Epub 2020 Dec 7. Proc Natl Acad Sci U S A. 2020. PMID: 33288725 Free PMC article.
References
- Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. - PubMed
- Laspia MF, Rice AP, Mathews MB. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell. 1989;59:283–292. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K99/R00 A112185/PHS HHS/United States
- AI29135/AI/NIAID NIH HHS/United States
- R01 AI029135/AI/NIAID NIH HHS/United States
- P50 GM082250-03/GM/NIGMS NIH HHS/United States
- P50 GM082250/GM/NIGMS NIH HHS/United States
- R01 AI029135-20/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials