Estimation of effect size distribution from genome-wide association studies and implications for future discoveries - PubMed (original) (raw)
Estimation of effect size distribution from genome-wide association studies and implications for future discoveries
Ju-Hyun Park et al. Nat Genet. 2010 Jul.
Abstract
We report a set of tools to estimate the number of susceptibility loci and the distribution of their effect sizes for a trait on the basis of discoveries from existing genome-wide association studies (GWASs). We propose statistical power calculations for future GWASs using estimated distributions of effect sizes. Using reported GWAS findings for height, Crohn's disease and breast, prostate and colorectal (BPC) cancers, we determine that each of these traits is likely to harbor additional loci within the spectrum of low-penetrance common variants. These loci, which can be identified from sufficiently powerful GWASs, together could explain at least 15-20% of the known heritability of these traits. However, for BPC cancers, which have modest familial aggregation, our analysis suggests that risk models based on common variants alone will have modest discriminatory power (63.5% area under curve), even with new discoveries.
Figures
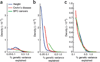
Figure 1
Nonparametric estimates for distributions of effect sizes for susceptibility loci. (a) Curves based only on observed susceptibility loci; these curves are distorted because loci with larger effect sizes are more likely to have been detected. (b) Curves based on estimated susceptibility loci, representative of the population of all susceptibility loci. (c) Estimated nonparametric distributions after normalization over the common observed range for the three traits.
Figure 2
Receiver operating characteristic curves for genetic risk models. (a,b) Curves for Crohn’s disease (a) and BPC cancers (b). AUC is a measure of the discriminatory power of the risk model. Blue, a theoretical genetic risk model that explains all of the known familial risk of the trait. Green, a risk model that includes all of the susceptibility loci (142 for Crohn’s disease and 67 on average for BPC cancers) estimated to exist within the range of effect sizes seen in the current GWASs. Red, a risk model that includes only known susceptibility loci (~30 for Crohn’s disease and ~7 on average for each of the BPC cancers), which we used to estimate the distribution of effect sizes of these traits. Black, reference line corresponding to a model without discriminatory power in which cases have the same distribution of risk as controls.
Similar articles
- Hints of hidden heritability in GWAS.
Gibson G. Gibson G. Nat Genet. 2010 Jul;42(7):558-60. doi: 10.1038/ng0710-558. Nat Genet. 2010. PMID: 20581876 - Strategies for developing prediction models from genome-wide association studies.
Wu J, Pfeiffer RM, Gail MH. Wu J, et al. Genet Epidemiol. 2013 Dec;37(8):768-77. doi: 10.1002/gepi.21762. Epub 2013 Oct 25. Genet Epidemiol. 2013. PMID: 24166696 - Common SNPs explain a large proportion of the heritability for human height.
Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME, Visscher PM. Yang J, et al. Nat Genet. 2010 Jul;42(7):565-9. doi: 10.1038/ng.608. Epub 2010 Jun 20. Nat Genet. 2010. PMID: 20562875 Free PMC article. - Genome-wide association studies in common cancers--what have we learnt?
Varghese JS, Easton DF. Varghese JS, et al. Curr Opin Genet Dev. 2010 Jun;20(3):201-9. doi: 10.1016/j.gde.2010.03.012. Epub 2010 Apr 24. Curr Opin Genet Dev. 2010. PMID: 20418093 Review. - Germline DNA variations in breast cancer predisposition and prognosis: a systematic review of the literature.
Sapkota Y. Sapkota Y. Cytogenet Genome Res. 2014;144(2):77-91. doi: 10.1159/000369045. Epub 2014 Nov 15. Cytogenet Genome Res. 2014. PMID: 25401968 Review.
Cited by
- Missing Causality and Heritability of Autoimmune Hepatitis.
Czaja AJ. Czaja AJ. Dig Dis Sci. 2023 Apr;68(4):1585-1604. doi: 10.1007/s10620-022-07728-w. Epub 2022 Oct 19. Dig Dis Sci. 2023. PMID: 36261672 - Associations between TRPV4 genotypes and body mass index in Taiwanese subjects.
Duan DM, Wu S, Hsu LA, Teng MS, Lin JF, Sun YC, Cheng CF, Ko YL. Duan DM, et al. Mol Genet Genomics. 2015 Aug;290(4):1357-65. doi: 10.1007/s00438-015-0996-8. Epub 2015 Feb 3. Mol Genet Genomics. 2015. PMID: 25647731 - Cancer heterogeneity: origins and implications for genetic association studies.
Urbach D, Lupien M, Karagas MR, Moore JH. Urbach D, et al. Trends Genet. 2012 Nov;28(11):538-43. doi: 10.1016/j.tig.2012.07.001. Epub 2012 Jul 31. Trends Genet. 2012. PMID: 22858414 Free PMC article. - Urinary bladder cancer susceptibility markers. What do we know about functional mechanisms?
Dudek AM, Grotenhuis AJ, Vermeulen SH, Kiemeney LA, Verhaegh GW. Dudek AM, et al. Int J Mol Sci. 2013 Jun 10;14(6):12346-66. doi: 10.3390/ijms140612346. Int J Mol Sci. 2013. PMID: 23752272 Free PMC article. Review. - Robust Huber-LASSO for improved prediction of protein, metabolite and gene expression levels relying on individual genotype data.
Deutelmoser H, Scherer D, Brenner H, Waldenberger M; INTERVAL study; Suhre K, Kastenmüller G, Lorenzo Bermejo J. Deutelmoser H, et al. Brief Bioinform. 2021 Jul 20;22(4):bbaa230. doi: 10.1093/bib/bbaa230. Brief Bioinform. 2021. PMID: 33063116 Free PMC article.
References
- Hirschhorn JN. Genomewide association studies–illuminating biologic pathways. N. Engl. J. Med. 2009;360:1699–1701. - PubMed
- Goldstein DB. Common genetic variation and human traits. N. Engl. J. Med. 2009;360:1696–1698. - PubMed
- Kraft P, et al. Beyond odds ratios–communicating disease risk based on genetic profiles. Nat. Rev. Genet. 2009;10:264–269. - PubMed
- Pharoah PD, et al. Polygenic susceptibility to breast cancer and implications for prevention. Nat. Genet. 2002;31:33–36. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources