The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer - PubMed (original) (raw)
The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer
Michelle L Stewart et al. Nat Chem Biol. 2010 Aug.
Abstract
The development of selective inhibitors for discrete anti-apoptotic BCL-2 family proteins implicated in pathologic cell survival remains a formidable but pressing challenge. Such precisely tailored compounds would serve as molecular probes and targeted therapies to study and treat human diseases driven by specific anti-apoptotic blockades. In particular, MCL-1 has emerged as a major resistance factor in human cancer. By screening a library of stabilized alpha-helix of BCL-2 domains (SAHBs), we determined that the MCL-1 BH3 helix is itself a potent and exclusive MCL-1 inhibitor. X-ray crystallography and mutagenesis studies defined key binding and specificity determinants, including the capacity to harness the hydrocarbon staple to optimize affinity while preserving selectivity. MCL-1 SAHB directly targets MCL-1, neutralizes its inhibitory interaction with pro-apoptotic BAK and sensitizes cancer cells to caspase-dependent apoptosis. By leveraging nature's solution to ligand selectivity, we generated an MCL-1-specific agent that defines the structural and functional features of targeted MCL-1 inhibition.
Figures
Figure 1
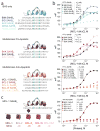
Identification of an MCL-1-selective BH3 domain. (a) A panel of Stabilized Alpha-Helix of BCL-2 domains (SAHBs) was designed based on the BH3 domains of pro- and anti-apoptotic BCL-2 family members. A pair of crosslinking non-natural amino acids (X) were substituted at the indicated i, i+4 position of the non-interacting helical surface and “stapled” by ruthenium-catalyzed olefin metathesis. To optimize the activity of the Grubbs’ ruthenium catalyst, sulfur-containing methionines were replaced with norleucines, which are designated by the letter B. (b) Dissociation constants for the binding of fluorescently labeled SAHBs to MCL-1ΔNΔC were determined by fluorescence polarization assay (FPA) and nonlinear regression analysis. (c) Among the SAHBs that bound MCL-1ΔNΔC with high affinity, only MCL-1 SAHB_A_ displayed a potent and exclusive interaction with MCL-1ΔNΔC, as evidenced by FPA performed with FITC-MCL-1 SAHB_A_ against a broad panel of anti-apoptotic targets. Data are mean and s.d. for experiments performed in at least triplicate.
Figure 2
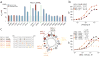
Binding and specificity determinants of the MCL-1 BH3 helix. (a) A panel of sequential alanine mutants (alanine scan) of FITC-MCL-1 SAHB_A_ was generated for FPA binding analysis, revealing key residues within the core BH3 sequence required for high affinity MCL-1ΔNΔC binding. Glutamate mutagenesis was also performed to evaluate the contribution of native alanine and glycine residues to MCL-1ΔNΔC binding. *, K_D_ >10 μM. (b) A single point mutation of V220F eliminated the MCL-1 specificity of MCL-1 SAHB_A_, conferring binding affinity to both MCL-1ΔNΔC and BCL-XLΔC, as demonstrated by FPA. (c) Sampling a variety of staple positions along the α-helical surface revealed disruption of MCL-1ΔNΔC binding only by the G217,Q221 staple (MCL-1 SAHB_C_), which is located at the hydrophobic binding interface. MCL-1 SAHB_D_ exhibited the strongest binding activity (K_D_, 10 nM), with 4-fold improvement over the parental MCL-1 SAHB_A_. Data are mean and s.d. for experiments performed in at least triplicate.
Figure 3
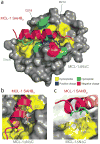
Crystal structure of the MCL-1 SAHB_D_/MCL-1ΔNΔC complex. (a) MCL-1 SAHB_D_ engages MCL-1ΔNΔC at the canonical BH3 binding groove of anti-apoptotic proteins, as determined by x-ray crystallography at 2.32-Å resolution (PDB 3MK8). Hydrophobic interactions at the binding interface are reinforced by a complementary polar interaction network that involves MCL-1 SAHB_D_ residues R214 and D218 and MCL-1ΔNΔC residues S255, D256, N260, and R263. The side chains of hydrophobic, positively charged, negatively charged and hydrophilic residues are colored yellow, blue, red and green, respectively. (b) The core BH3 residues L213, V216, G217 and V220 of MCL-1 SAHB_D_ make direct contact with a hydrophobic cleft at the surface of MCL- 1ΔNΔC. (c) The hydrocarbon staple, bearing an olefin in the cis conformation, contributes additional hydrophobic contacts at the perimeter of the core interaction site.
Figure 4
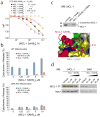
MCL-1 SAHB_D_ dissociates the inhibitory MCL-1/BAK complex in vitro and in situ, and sensitizes BAK-dependent mitochondrial cytochrome c release. (a) MCL-1 SAHBs effectively prevent sequestration of the BAK BH3 helix by MCL-1ΔNΔC, as demonstrated by competition FPA. N.D., no detected displacement. (b) MCL-1 SAHB_D_ dose-responsively sensitized BID BH3-induced and BAK-dependent mitochondrial apoptosis, as measured by cytochrome c release assay performed on wild type and Bak_−/− mitochondria. (c) An OPM2 multiple myeloma cellular lysate was incubated with the indicated biotinylated MCL-1 SAHB_D constructs in the presence of ultraviolet light, followed by streptavidin-based purification, stringent washing to remove non-covalent binders, elution, and MCL-1 western analysis. The photoreactive MCL-1 pSAHB_D_, generated by replacing L210 with a benzophenone-bearing non-natural amino acid (Bpa), directly crosslinked to native MCL-1 within the cellular lysate, whereas no covalent crosslinking was observed for MCL-1 SAHB_D_, which lacked the photoreactive benzophenone moiety. (d) The native interaction between BAK and MCL-1 was dose-responsively disrupted by treatment of OPM2 cells with MCL-1 SAHB_D_, as assessed by MCL-1 immunoprecipitation and BAK western analysis. Binding and cytochrome c release data are mean and s.d. for experiments performed in at least triplicate. Vehicle, deionized water.
Figure 5
Selective MCL-1 targeting by MCL-1 SAHB_D_ sensitizes death receptor signaling and induces caspase-dependent cancer cell apoptosis. (a) Jurkat T-cell leukemia and (b) OPM2 cells were exposed to MCL-1 SAHB_D_ singly and in combination with low dose death receptor agonists TRAIL and Fas ligand (FasL) in the presence or absence of the pan-caspase inhibitor, z-VAD. Cell viability measured by MTT assay at 24 hours revealed dose-responsive and caspase-dependent apoptosis sensitization of Jurkat (TRAIL and FasL) and OPM2 (TRAIL) cells by MCL-1 SAHB_D_. The capacity of MCL-1 SAHB_D_ to sensitize (c) Jurkat and (d) OPM2 cells to death receptor stimuli correlated with dose-responsive activation of caspase 3/7, as measured by luminescence of DEVD-cleaved substrate. Data are mean and s.d. for experiments performed in at least triplicate. Vehicle, deionized water.
Comment in
- Stapled peptides: Magic bullets in nature's arsenal.
Kritzer JA. Kritzer JA. Nat Chem Biol. 2010 Aug;6(8):566-7. doi: 10.1038/nchembio.407. Nat Chem Biol. 2010. PMID: 20644540 No abstract available.
Similar articles
- Specific cleavage of Mcl-1 by caspase-3 in tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis in Jurkat leukemia T cells.
Weng C, Li Y, Xu D, Shi Y, Tang H. Weng C, et al. J Biol Chem. 2005 Mar 18;280(11):10491-500. doi: 10.1074/jbc.M412819200. Epub 2005 Jan 6. J Biol Chem. 2005. PMID: 15637055 - Mcl-1 interacts with truncated Bid and inhibits its induction of cytochrome c release and its role in receptor-mediated apoptosis.
Clohessy JG, Zhuang J, de Boer J, Gil-Gómez G, Brady HJ. Clohessy JG, et al. J Biol Chem. 2006 Mar 3;281(9):5750-9. doi: 10.1074/jbc.M505688200. Epub 2005 Dec 27. J Biol Chem. 2006. PMID: 16380381 - Solution structure of prosurvival Mcl-1 and characterization of its binding by proapoptotic BH3-only ligands.
Day CL, Chen L, Richardson SJ, Harrison PJ, Huang DC, Hinds MG. Day CL, et al. J Biol Chem. 2005 Feb 11;280(6):4738-44. doi: 10.1074/jbc.M411434200. Epub 2004 Nov 18. J Biol Chem. 2005. PMID: 15550399 - A novel BH3 mimetic efficiently induces apoptosis in melanoma cells through direct binding to anti-apoptotic Bcl-2 family proteins, including phosphorylated Mcl-1.
Liu Y, Xie M, Song T, Sheng H, Yu X, Zhang Z. Liu Y, et al. Pigment Cell Melanoma Res. 2015 Mar;28(2):161-70. doi: 10.1111/pcmr.12325. Epub 2014 Dec 18. Pigment Cell Melanoma Res. 2015. PMID: 25324174 - Structural biology of the Bcl-2 family of proteins.
Petros AM, Olejniczak ET, Fesik SW. Petros AM, et al. Biochim Biophys Acta. 2004 Mar 1;1644(2-3):83-94. doi: 10.1016/j.bbamcr.2003.08.012. Biochim Biophys Acta. 2004. PMID: 14996493 Review.
Cited by
- Challenges in Targeting a Basic Helix-Loop-Helix Transcription Factor with Hydrocarbon-Stapled Peptides.
Edwards AL, Meijer DH, Guerra RM, Molenaar RJ, Alberta JA, Bernal F, Bird GH, Stiles CD, Walensky LD. Edwards AL, et al. ACS Chem Biol. 2016 Nov 18;11(11):3146-3153. doi: 10.1021/acschembio.6b00465. Epub 2016 Oct 4. ACS Chem Biol. 2016. PMID: 27643505 Free PMC article. - Cellular Uptake and Ultrastructural Localization Underlie the Pro-apoptotic Activity of a Hydrocarbon-stapled BIM BH3 Peptide.
Edwards AL, Wachter F, Lammert M, Huhn AJ, Luccarelli J, Bird GH, Walensky LD. Edwards AL, et al. ACS Chem Biol. 2015 Sep 18;10(9):2149-57. doi: 10.1021/acschembio.5b00214. Epub 2015 Jul 21. ACS Chem Biol. 2015. PMID: 26151238 Free PMC article. - Targeting MCL-1 in cancer: current status and perspectives.
Wang H, Guo M, Wei H, Chen Y. Wang H, et al. J Hematol Oncol. 2021 Apr 21;14(1):67. doi: 10.1186/s13045-021-01079-1. J Hematol Oncol. 2021. PMID: 33883020 Free PMC article. Review. - Peptide bicycles that inhibit the Grb2 SH2 domain.
Quartararo JS, Wu P, Kritzer JA. Quartararo JS, et al. Chembiochem. 2012 Jul 9;13(10):1490-6. doi: 10.1002/cbic.201200175. Epub 2012 Jun 11. Chembiochem. 2012. PMID: 22689355 Free PMC article. - Discovery of a Myeloid Cell Leukemia 1 (Mcl-1) Inhibitor That Demonstrates Potent In Vivo Activities in Mouse Models of Hematological and Solid Tumors.
Tarr JC, Salovich JM, Aichinger M, Jeon K, Veerasamy N, Sensintaffar JL, Arnhof H, Samwer M, Christov PP, Kim K, Wunberg T, Schweifer N, Trapani F, Arnold A, Martin F, Zhao B, Miriyala N, Sgubin D, Fogarty S, Moore WJ, Stott GM, Olejniczak ET, Engelhardt H, Rudolph D, Lee T, McConnell DB, Fesik SW. Tarr JC, et al. J Med Chem. 2024 Aug 22;67(16):14370-14393. doi: 10.1021/acs.jmedchem.4c01188. Epub 2024 Aug 5. J Med Chem. 2024. PMID: 39102508 Free PMC article.
References
- Tsujimoto Y, Cossman J, Jaffe E, Croce CM. Involvement of the bcl-2 gene in human follicular lymphoma. Science. 1985;228:1440–3. - PubMed
- Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–19. - PubMed
- Sattler M, et al. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science. 1997;275:983–6. - PubMed
- Muchmore SW, et al. X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death. Nature. 1996;381:335–41. - PubMed
- Chen L, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P41 RR015301/RR/NCRR NIH HHS/United States
- 5P01CA92625/CA/NCI NIH HHS/United States
- P01 CA092625/CA/NCI NIH HHS/United States
- RR-15301/RR/NCRR NIH HHS/United States
- P01 CA092625-09/CA/NCI NIH HHS/United States
- P01 CA092625-09S1/CA/NCI NIH HHS/United States
- 5R01GM084181/GM/NIGMS NIH HHS/United States
- P01 CA092625-08/CA/NCI NIH HHS/United States
- R01 GM084181-03/GM/NIGMS NIH HHS/United States
- R01 GM084181-04/GM/NIGMS NIH HHS/United States
- R01 GM084181/GM/NIGMS NIH HHS/United States
- F31 CA144566/CA/NCI NIH HHS/United States
- 1F31CA144566/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases