Requirement of the histone demethylase LSD1 in Snai1-mediated transcriptional repression during epithelial-mesenchymal transition - PubMed (original) (raw)
Requirement of the histone demethylase LSD1 in Snai1-mediated transcriptional repression during epithelial-mesenchymal transition
T Lin et al. Oncogene. 2010.
Abstract
Epithelial-mesenchymal transition (EMT) has pivotal roles during embryonic development and carcinoma progression. Members of the Snai1 family of zinc finger transcription factors are central mediators of EMT and induce EMT in part by directly repressing epithelial markers such as E-cadherin, a gatekeeper of the epithelial phenotype and a suppressor of tumor invasion. However, the molecular mechanism underlying Snai1-mediated transcriptional repression remains incompletely understood. Here we show that Snai1 physically interacts with and recruits the histone demethylase LSD1 (KDM1A) to epithelial gene promoters. LSD1 removes dimethylation of lysine 4 on histone H3 (H3K4m2), a covalent histone modification associated with active chromatin. Importantly, LSD1 is essential for Snai1-mediated transcriptional repression and for maintenance of the silenced state of Snai1 target genes in invasive cancer cells. In the absence of LSD1, Snai1 fails to repress E-cadherin. In cancer cells in which E-cadherin is silenced, depletion of LSD1 results in partial de-repression of epithelial genes and elevated H3K4m2 levels at the E-cadherin promoter. These results underline the critical role of LSD1 in Snai1-dependent transcriptional repression of epithelial markers and suggest that the LSD1 complex could be a potential therapeutic target for prevention of EMT-associated tumor invasion.
Conflict of interest statement
Conflict of Interest:
The authors declare no conflict of interest.
Figures
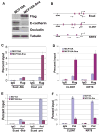
Figure 1. Snai1 directly represses epithelial genes
(A) Ectopic expression of Flag-tagged Snai1 in MCF10A cells downregulates epithelial markers E-cadherin and Occludin. Protein lysates from control and Snai1-expressing MCF10A cells were probed by Western blotting with indicated antibodies. (B) Diagrams of the proximal promoters of E-cadherin, CLDN7, and KRT8. Vertical bars represent E-boxes. Arrows indicate primers used in ChIP assays. (C) Snai1 is specifically enriched at the proximal promoter of E-cadherin in vivo as demonstrated by ChIP analysis. The results represent percentage of input chromatin and error bars indicate standard deviations (S.D.) from triplicate experiments. (D) Snai1 is present at the promoters of CLDN7 and KRT8 in Snai1-expressing cells. (E) Expression of Snai1 in MCF10A dissociates RNA polymerase II (Pol) from the E-cadherin promoter. (F) Snai1 decreases Pol II binding to the CLDN7 and KRT8 promoters.
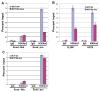
Figure 2. Snai1 reduces H3K4 dimethylation at its target gene promoters
(A) Snai1 expression causes a reduction in H3K4m2 levels at the E-cadherin promoter, but not in the 5kb upstream region, as determined by ChIP analysis. (B) Snai1 reduces H3K4m2 levels at the CLDN7 and KRT8 promoters. (C) The H3K4m3 mark at the E-cadherin promoter is not affected by Snai1.
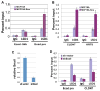
Figure 3. Snai1 physically interacts with LSD1 in vitro and in vivo
(A) Schematic diagram of SNAG-GST and GST-ZF fusion proteins used in GST pull-down assays. (B) The SNAG domain is sufficient for association with the LSD1 complex. Whole cell lysate prepared from HEK293 cells transfected with Flag-LSD1 was incubated with GST and the SNAG-GST fusion protein and followed by Western blot analysis using anti-Flag, anti-CoREST, and anti-HDAC1 antibodies. Coomassie staining shows the protein loading of GST and SNAG-GST. (C) LSD1 directly interacts with the SNAG domain. 35S-labeled LSD1 protein was mixed with GST, SNAG-GST, or GST-ZF fusion proteins. Bound LSD1 was then detected by autoradiography after SDS/PAGE. Coomassie staining of GST proteins was shown. (D) The amino-terminus of LSD1 interacts with SNAG. (E) Exogenous Snai1 associates with endogenous LSD1 in intact cells. Co-immunoprecipitation of whole cell lysate from HEK293 cells overexpressing Flag-tagged wild-type Snai1 or Snai1-P2A mutant was performed with anti-Flag antibody. The presence of LSD1 in the precipitates was shown by Western blotting with an anti-LSD1 antibody. Anti-Flag Western blotting indicates expression of wild-type and mutant Snai1. (F) Endogenous Snai1 and LSD1 form a complex in vivo. MDA-MB-231 cells were lysed and incubated with two anti-Snai1 antibodies (#1 fromSantaCruz; #2 from Cellsignaling) or control IgG, followed by Western blotting with the LSD1 antibody.
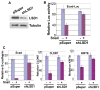
Figure 4. LSD1 is recruited to epithelial gene promoters by Snai1
(A) Occupancy of LSD1 at the E-cadherin promoter is increased in Snai1-expressing cells compared to control MCF10A cells as shown by ChIP analysis with the LSD1 antibody. (B) The binding of LSD1 to the CLDN7 and KRT8 promoters is increased in MCF10A cells expressing Snai1. (C) Quantitative RT-PCR analysis shows efficient depletion of Snai1 in MDA-MB-231 cells by a retroviral shRNA. (D) The presence of LSD1 at the E-cadherin and CLDN7 promoters is reduced in MDA-MB-231 cells depleted of Snai1.
Figure 5. LSD1 is essential for Snai1-mediated repression
(A) Verification of LSD1 knockdown by Western blotting. MCF7 cells were infected with retroviral empty vector pSuper or with shRNA targeting LSD1, followed by Western blotting assays. (B) Depletion of LSD1 impairs the repressive activity of Snai1 in reporter-based assays. Snai1 fails to repress a luciferase reporter driven by the E-cadherin promoter in LSD1-depleted cells. Error bars indicate S.D. from three independent experiments. (C) LSD1 is essential for Snai1-mediated repression of endogenous epithelial genes. Expression of E-cadherin, CLDN7, and KRT8 was determined by quantitative RT-PCR.
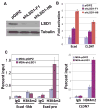
Figure 6. LSD1 is required to maintain the silenced status of Snai1 target genes in invasive cancer cells
(A) Validation of LSD1 depletion in MDA-MB-231 cells with two lentiviral shRNAs by Western blotting with denoted antibodies. Tubulin serves as a loading control. (B) Expression of E-cadherin and CLDN7 is upregulated in MDA-MB-231 cells depleted of LSD1. The RNA levels of the two genes were normalized against GAPDH by quantitative RT-PCR analysis. (C) LSD1 depletion in MDA-MB-231 cells increases H3K4m2 levels specifically at the E-cadherin and CLDN7 promoters, as determined by ChIP assays.
Similar articles
- The malignant brain tumor (MBT) domain protein SFMBT1 is an integral histone reader subunit of the LSD1 demethylase complex for chromatin association and epithelial-to-mesenchymal transition.
Tang M, Shen H, Jin Y, Lin T, Cai Q, Pinard MA, Biswas S, Tran Q, Li G, Shenoy AK, Tongdee E, Lin S, Gu Y, Law BK, Zhou L, Mckenna R, Wu L, Lu J. Tang M, et al. J Biol Chem. 2013 Sep 20;288(38):27680-27691. doi: 10.1074/jbc.M113.482349. Epub 2013 Aug 8. J Biol Chem. 2013. PMID: 23928305 Free PMC article. - TdIF1-LSD1 Axis Regulates Epithelial-Mesenchymal Transition and Metastasis via Histone Demethylation of E-Cadherin Promoter in Lung Cancer.
Liu Q, Xiong J, Xu D, Hao N, Zhang Y, Sang Y, Wang Z, Zheng X, Min J, Diao H, Raphael J, Maleki Vareki S, Koropatnick J, Min W. Liu Q, et al. Int J Mol Sci. 2021 Dec 27;23(1):250. doi: 10.3390/ijms23010250. Int J Mol Sci. 2021. PMID: 35008676 Free PMC article. - HMG20A is required for SNAI1-mediated epithelial to mesenchymal transition.
Rivero S, Ceballos-Chávez M, Bhattacharya SS, Reyes JC. Rivero S, et al. Oncogene. 2015 Oct 8;34(41):5264-76. doi: 10.1038/onc.2014.446. Epub 2015 Feb 2. Oncogene. 2015. PMID: 25639869 - Epigenetic regulation of epithelial to mesenchymal transition by the Lysine-specific demethylase LSD1/KDM1A.
Ambrosio S, Saccà CD, Majello B. Ambrosio S, et al. Biochim Biophys Acta Gene Regul Mech. 2017 Sep;1860(9):905-910. doi: 10.1016/j.bbagrm.2017.07.001. Epub 2017 Jul 15. Biochim Biophys Acta Gene Regul Mech. 2017. PMID: 28720390 Review. - Histone demethylase LSD1 controls the phenotypic plasticity of cancer cells.
Hino S, Kohrogi K, Nakao M. Hino S, et al. Cancer Sci. 2016 Sep;107(9):1187-92. doi: 10.1111/cas.13004. Epub 2016 Sep 1. Cancer Sci. 2016. PMID: 27375009 Free PMC article. Review.
Cited by
- A Novel Class I HDAC Inhibitor, AW01178, Inhibits Epithelial-Mesenchymal Transition and Metastasis of Breast Cancer.
Liu X, Chen Y, Li Y, Shen Y, Dong S, Tan J. Liu X, et al. Int J Mol Sci. 2024 Jun 30;25(13):7234. doi: 10.3390/ijms25137234. Int J Mol Sci. 2024. PMID: 39000339 Free PMC article. - Tranylcypromine, a lysine-specific demethylase 1 (LSD1) inhibitor, suppresses lesion growth and improves generalized hyperalgesia in mouse with induced endometriosis.
Sun Q, Ding D, Liu X, Guo SW. Sun Q, et al. Reprod Biol Endocrinol. 2016 Apr 9;14:17. doi: 10.1186/s12958-016-0154-0. Reprod Biol Endocrinol. 2016. PMID: 27062244 Free PMC article. - KDM1A promotes thyroid cancer progression and maintains stemness through the Wnt/β-catenin signaling pathway.
Zhang W, Ruan X, Li Y, Zhi J, Hu L, Hou X, Shi X, Wang X, Wang J, Ma W, Gu P, Zheng X, Gao M. Zhang W, et al. Theranostics. 2022 Jan 3;12(4):1500-1517. doi: 10.7150/thno.66142. eCollection 2022. Theranostics. 2022. PMID: 35198054 Free PMC article. - Targeting the LSD1/KDM1 Family of Lysine Demethylases in Cancer and Other Human Diseases.
Mao F, Shi YG. Mao F, et al. Adv Exp Med Biol. 2023;1433:15-49. doi: 10.1007/978-3-031-38176-8_2. Adv Exp Med Biol. 2023. PMID: 37751134 Review. - Interactions and Feedbacks in E-Cadherin Transcriptional Regulation.
Ramirez Moreno M, Stempor PA, Bulgakova NA. Ramirez Moreno M, et al. Front Cell Dev Biol. 2021 Jun 28;9:701175. doi: 10.3389/fcell.2021.701175. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34262912 Free PMC article.
References
- Batlle E, Sancho E, Francí C, Domínguez D, Monfar M, Baulida J, et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. - PubMed
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
- Cano A, Pérez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. - PubMed
- De Craene B, Gilbert B, Stove C, Bruyneel E, van Roy F, Berx G. The transcription factor snail induces tumor cell invasion through modulation of the epithelial cell differentiation program. Cancer Res. 2005;65:6237–6244. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA137021/CA/NCI NIH HHS/United States
- R01 CA137021-01A1/CA/NCI NIH HHS/United States
- R01 CA137021-02/CA/NCI NIH HHS/United States
- R01CA137021/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials