Multiple functions of Ldb1 required for beta-globin activation during erythroid differentiation - PubMed (original) (raw)
Multiple functions of Ldb1 required for beta-globin activation during erythroid differentiation
Sang-Hyun Song et al. Blood. 2010.
Abstract
Ldb1 and erythroid partners SCL, GATA-1, and LMO2 form a complex that is required to establish spatial proximity between the β-globin locus control region and gene and for transcription activation during erythroid differentiation. Here we show that Ldb1 controls gene expression at multiple levels. Ldb1 stabilizes its erythroid complex partners on β-globin chromatin, even though it is not one of the DNA-binding components. In addition, Ldb1 is necessary for enrichment of key transcriptional components in the locus, including P-TEFb, which phosphorylates Ser2 of the RNA polymerase C-terminal domain for efficient elongation. Furthermore, reduction of Ldb1 results in the inability of the locus to migrate away from the nuclear periphery, which is necessary to achieve robust transcription of β-globin in nuclear transcription factories. Ldb1 contributes these critical functions at both embryonic and adult stages of globin gene expression. These results implicate Ldb1 as a factor that facilitates nuclear relocation for transcription activation.
Figures
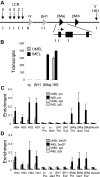
Figure 1
Recruitment of elongation competent Ser2P RNA pol II to the β-globin locus during mouse MEL cell differentiation. (A) The mouse β-globin locus is diagrammed, and the positions of TaqMan probes used for real-time PCR are indicated below and named on the graphs. (B) MEL cells were treated with 2% DMSO for 4 days (IMEL) or without DMSO (UMEL), and globin mRNA was analyzed by quantitative real-time RT-PCR. Data are mean ± SEM, and 18S ribosomal RNA transcripts served as a control. (C-D) ChIP was performed MEL cell chromatin and antibodies to (C) RNA pol II or (D) Ser2P pol II. Three different chromatin preparations were analyzed by quantitative PCR. Error bars represent the SEM. Necdin, not transcribed in these cells, served as a negative control.
Figure 2
Enrichment of P-TEFb kinase at the β-globin LCR and promoter region in differentiating MEL cells and alterations in H3ac and H3K4me3. (A-C) Chromatin was prepared from MEL cells before and after 4 days of 2% DMSO treatment. ChIP was performed with antibodies to (A) Cdk9 or (B) CycT1. Necdin served as a negative control. Data are mean ± SEM. (C) Antibodies to acH3 or H3K4me3 were used in ChIP and graphed together by setting the highest value obtained with each antibody equal to 1. Lines represent AcH3; and bars, H3K4me3.
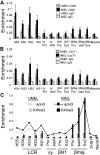
Figure 3
Ldb1 knockdown inhibits enrichment of the Ldb1 complex and Ser2P RNA pol II at the β-globin promoter and LCR in MEL cells. (A) MEL cells were stably transduced with Ldb1 shRNA or with an empty virus (Ctrl) and individual clones were isolated. Reduced Ldb1 expression in 2 stable Ldb1 knockdown clones (KD1 and KD2) was confirmed by Western blot analysis before (−) and after 4 days of 2% DMSO treatment (+). GATA-1 and α-tubulin served as positive and internal controls, respectively. (B) Stable clones were treated with 2% DMSO. Total RNA was isolated at day 4, and β-globin expression was analyzed by quantitative real-time RT-PCR. Each value was normalized with 18S ribosomal RNA. Three RNA preparations were analyzed. Error bars represent the SEM. (C) ChIP was carried out using KD1 cells after 4 days of 2% DMSO induction using antibodies to acH3 and H3K4me3, and the data were graphed together by setting the highest level for each antibody equal to 1. (D) The ChIP assay was performed with KD1 cells after 4 days of 2% DMSO induction using antibodies to Ldb1, GATA-1, LMO2, and SCL. Error bars represent the SEM for multiple independent chromatin preparations. (E-F) ChIP was carried out as for panel D using antibodies to Ser2P pol II (E) or Cdk9 (F). Necdin served as a negative control. Error bars represent the SEM for multiple independent chromatin preparations. ChIP assays with KD2 cells gave similar results (not shown) to those in panels C through F.
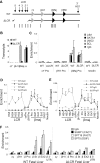
Figure 4
The Ldb1 complex and Ser2P RNA pol II are enriched at the β-globin promoter in fetal liver erythroid cells of mice homozygous for a deletion of the LCR. (A) The mouse β-globin locus is diagrammed, and the positions of TaqMan probes used for real-time PCR are indicated below and named on the graphs. Endogenous (WT) and LCR-deleted (ΔLCR) murine β-globin loci are depicted. (B) Total RNA was isolated from E14.5 fetal liver of WT or ΔLCR mice, and globin expression was analyzed by quantitative real-time RT-PCR. α-Globin was used as a positive control of RNA expression. Each value was normalized with 18S ribosomal RNA. Error bars represent the SEM for several independent RNA preparations. (C-F) Chromatin was prepared from E14.5 fetal liver of WT (+) or ΔLCR (−) mice, and then ChIP and quantitative PCR were performed with (C) Ldb1, GATA-1, LMO2, and SCL, (D) Ser2P pol II, and (E) Cdk9, or (F) DSIF and FACT components as indicated on the graphs. Necdin served as a negative control. Error bars represent the SEM among independent chromatin preparations.
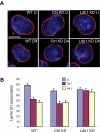
Figure 5
Ldb1 is required for localization of the β-globin locus away from the nuclear periphery during differentiation. (A) Three-dimensional immuno-FISH analysis of untreated cells and cells treated with DMSO. WT indicates MEL cells; Ctrl KD, MEL cells with control shRNA; KD, MEL cells with shRNA directed to Ldb1; U, uninduced; and 4D, treated with DMSO for 4 days. Red represents nuclear lamin immunofluorescence; and green, probe detecting the β-globin locus. Bar represents 5 μm. For details see “Image acquisition and analysis.” (B) Quantitation of association of globin loci with the nuclear lamina before and after DMSO treatment for 3 days (3d) or 4 days (4d). More than 100 cells were scored for each determination. Error bars represent the SE.
Figure 6
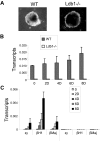
Importance of Ldb1 to embryonic and adult β-globin gene expression during erythroid differentiation. Wild-type (WT) or Ldb1-null mutant (Ldb1−/−) ES cells were differentiated along erythroid lines into EBs with erythropoietin for 8 days as described under “EB differentiation.” (A) Morphology of WT or Ldb1−/− EB after 8 days of differentiation. (B-C) Total RNA was isolated at the indicated day of differentiation to determine expression of (B) Ldb1, or (C) ϵγ, βH1, and βmaj-globin. Each value was normalized with 18S ribosomal RNA. Error bars represent the SEM from independent RNA preparations.
Figure 7
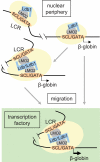
A model of dynamic long-range communication between the β-globin LCR and gene. Association of the Ldb1 complex (GATA-1/SCL/LMO2) and other activation components (shaded ovals), whose recruitment might be dependent on Ldb1, occupying the LCR and β-globin gene. Previous work comparing induced and uninduced MEL cells suggests that the Ldb1 complex binds first to the LCR. In the absence of the LCR, these components can co-occupy the β-globin gene, implying that promoter occupancy may normally occur independently; however, such occupancy is insufficient for high-level transcription. Full occupancy may be sufficient to establish LCR/β-globin proximity, before nuclear migration, via unknown mechanisms, and TF occupancy. Alternatively, the locus might migrate, again by unknown mechanisms, and establish LCR/β-globin proximity as a result of positioning within a TF. Proximity would then be stabilized by protein-protein interactions, possibly dependent on Ldb1.
Similar articles
- The Hematopoietic Stem and Progenitor Cell Cistrome: GATA Factor-Dependent cis-Regulatory Mechanisms.
Hewitt KJ, Johnson KD, Gao X, Keles S, Bresnick EH. Hewitt KJ, et al. Curr Top Dev Biol. 2016;118:45-76. doi: 10.1016/bs.ctdb.2016.01.002. Epub 2016 Feb 26. Curr Top Dev Biol. 2016. PMID: 27137654 Free PMC article. Review. - A positive role for NLI/Ldb1 in long-range beta-globin locus control region function.
Song SH, Hou C, Dean A. Song SH, et al. Mol Cell. 2007 Dec 14;28(5):810-22. doi: 10.1016/j.molcel.2007.09.025. Mol Cell. 2007. PMID: 18082606 Free PMC article. - The LIM-domain binding protein Ldb1 and its partner LMO2 act as negative regulators of erythroid differentiation.
Visvader JE, Mao X, Fujiwara Y, Hahm K, Orkin SH. Visvader JE, et al. Proc Natl Acad Sci U S A. 1997 Dec 9;94(25):13707-12. doi: 10.1073/pnas.94.25.13707. Proc Natl Acad Sci U S A. 1997. PMID: 9391090 Free PMC article. - Structure of the leukemia oncogene LMO2: implications for the assembly of a hematopoietic transcription factor complex.
El Omari K, Hoosdally SJ, Tuladhar K, Karia D, Vyas P, Patient R, Porcher C, Mancini EJ. El Omari K, et al. Blood. 2011 Feb 17;117(7):2146-56. doi: 10.1182/blood-2010-07-293357. Epub 2010 Nov 12. Blood. 2011. PMID: 21076045 - Ldb1 complexes: the new master regulators of erythroid gene transcription.
Love PE, Warzecha C, Li L. Love PE, et al. Trends Genet. 2014 Jan;30(1):1-9. doi: 10.1016/j.tig.2013.10.001. Epub 2013 Nov 27. Trends Genet. 2014. PMID: 24290192 Free PMC article. Review.
Cited by
- The Hematopoietic Stem and Progenitor Cell Cistrome: GATA Factor-Dependent cis-Regulatory Mechanisms.
Hewitt KJ, Johnson KD, Gao X, Keles S, Bresnick EH. Hewitt KJ, et al. Curr Top Dev Biol. 2016;118:45-76. doi: 10.1016/bs.ctdb.2016.01.002. Epub 2016 Feb 26. Curr Top Dev Biol. 2016. PMID: 27137654 Free PMC article. Review. - Functional interactions between erythroid Krüppel-like factor (EKLF/KLF1) and protein phosphatase PPM1B/PP2Cβ.
Yien YY, Bieker JJ. Yien YY, et al. J Biol Chem. 2012 May 4;287(19):15193-204. doi: 10.1074/jbc.M112.350496. Epub 2012 Mar 5. J Biol Chem. 2012. PMID: 22393050 Free PMC article. - Transcription regulation by distal enhancers: who's in the loop?
Stadhouders R, van den Heuvel A, Kolovos P, Jorna R, Leslie K, Grosveld F, Soler E. Stadhouders R, et al. Transcription. 2012 Jul-Aug;3(4):181-6. doi: 10.4161/trns.20720. Epub 2012 Jul 1. Transcription. 2012. PMID: 22771987 Free PMC article. Review. - Role of LDB1 in the transition from chromatin looping to transcription activation.
Krivega I, Dale RK, Dean A. Krivega I, et al. Genes Dev. 2014 Jun 15;28(12):1278-90. doi: 10.1101/gad.239749.114. Epub 2014 May 29. Genes Dev. 2014. PMID: 24874989 Free PMC article. - Dynamic enhancer-gene body contacts during transcription elongation.
Lee K, Hsiung CC, Huang P, Raj A, Blobel GA. Lee K, et al. Genes Dev. 2015 Oct 1;29(19):1992-7. doi: 10.1101/gad.255265.114. Genes Dev. 2015. PMID: 26443845 Free PMC article.
References
- Carter D, Chakalova L, Osborne CS, Dai Y, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32(4):623–626. - PubMed
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol Cell. 2002;10(6):1453–1465. - PubMed
- Dean A. On a chromosome far, far away: LCRs and gene regulation. Trends Genet. 2006;22:38–45. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DK44746/DK/NIDDK NIH HHS/United States
- R01 HL065440/HL/NHLBI NIH HHS/United States
- HL65440/HL/NHLBI NIH HHS/United States
- R37 DK044746/DK/NIDDK NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous