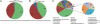Composition, variability, and temporal stability of the intestinal microbiota of the elderly - PubMed (original) (raw)
. 2011 Mar 15;108 Suppl 1(Suppl 1):4586-91.
doi: 10.1073/pnas.1000097107. Epub 2010 Jun 22.
Siobhán Cusack, Orla O'Sullivan, Rachel Greene-Diniz, Heleen de Weerd, Edel Flannery, Julian R Marchesi, Daniel Falush, Timothy Dinan, Gerald Fitzgerald, Catherine Stanton, Douwe van Sinderen, Michael O'Connor, Norma Harnedy, Kieran O'Connor, Colm Henry, Denis O'Mahony, Anthony P Fitzgerald, Fergus Shanahan, Cillian Twomey, Colin Hill, R Paul Ross, Paul W O'Toole
Affiliations
- PMID: 20571116
- PMCID: PMC3063589
- DOI: 10.1073/pnas.1000097107
Composition, variability, and temporal stability of the intestinal microbiota of the elderly
Marcus J Claesson et al. Proc Natl Acad Sci U S A. 2011.
Abstract
Alterations in the human intestinal microbiota are linked to conditions including inflammatory bowel disease, irritable bowel syndrome, and obesity. The microbiota also undergoes substantial changes at the extremes of life, in infants and older people, the ramifications of which are still being explored. We applied pyrosequencing of over 40,000 16S rRNA gene V4 region amplicons per subject to characterize the fecal microbiota in 161 subjects aged 65 y and older and 9 younger control subjects. The microbiota of each individual subject constituted a unique profile that was separable from all others. In 68% of the individuals, the microbiota was dominated by phylum Bacteroides, with an average proportion of 57% across all 161 baseline samples. Phylum Firmicutes had an average proportion of 40%. The proportions of some phyla and genera associated with disease or health also varied dramatically, including Proteobacteria, Actinobacteria, and Faecalibacteria. The core microbiota of elderly subjects was distinct from that previously established for younger adults, with a greater proportion of Bacteroides spp. and distinct abundance patterns of Clostridium groups. Analyses of 26 fecal microbiota datasets from 3-month follow-up samples indicated that in 85% of the subjects, the microbiota composition was more like the corresponding time-0 sample than any other dataset. We conclude that the fecal microbiota of the elderly shows temporal stability over limited time in the majority of subjects but is characterized by unusual phylum proportions and extreme variability.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Aggregate microbiota composition at phylum level in (A) 187 (161 time-0 and 26 time 3-mo) fecal samples from elderly subjects, (B) 9 healthy younger adult controls, and (C) at genus level in the 187 fecal samples from elderly subjects (the smaller pie chart to right is inserted for clarity). Only major taxonomic groups are shown; these cover 95% of all reads assigned to genus level.
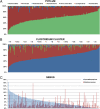
Fig. 2.
Interindividual variation in the proportion of major phyla (A), Clostridium clusters (B), and the genera Faecalibacterium and Ruminococcus (C) in 187 fecal samples from elderly subjects. Only the seven largest phyla are shown in A. Samples are ordered, from left, according to their Bacteroidetes proportion (A), cluster IV proportion of all reads assigned to Clostridium clusters (B), and Faecalibacterium proportion (C) to illustrate the dramatic interindividual variation.
Fig. 3.
(A) The core fecal microbiota of elderly subjects at the levels of phylum, genus and Clostridium cluster compared with similarly defined core microbiota from four studies of younger adults and nine young adult controls. (B) Unique microbiota defined by comparing elderly and younger adults (published datasets). (C) Unique microbiota defined by comparing elderly and younger adults (this study).
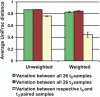
Fig. 4.
Variation in the microbiota between time-0 and time 3-mo samples from 26 subjects. Mean (± SEM) unweighted and weighted UniFrac distances between the samples as indicated.
Similar articles
- Intestinal microbiota in healthy adults: temporal analysis reveals individual and common core and relation to intestinal symptoms.
Jalanka-Tuovinen J, Salonen A, Nikkilä J, Immonen O, Kekkonen R, Lahti L, Palva A, de Vos WM. Jalanka-Tuovinen J, et al. PLoS One. 2011;6(7):e23035. doi: 10.1371/journal.pone.0023035. Epub 2011 Jul 28. PLoS One. 2011. PMID: 21829582 Free PMC article. Clinical Trial. - Characterization of the fecal microbiota using high-throughput sequencing reveals a stable microbial community during storage.
Carroll IM, Ringel-Kulka T, Siddle JP, Klaenhammer TR, Ringel Y. Carroll IM, et al. PLoS One. 2012;7(10):e46953. doi: 10.1371/journal.pone.0046953. Epub 2012 Oct 5. PLoS One. 2012. PMID: 23071673 Free PMC article. - Comparative analysis of Korean human gut microbiota by barcoded pyrosequencing.
Nam YD, Jung MJ, Roh SW, Kim MS, Bae JW. Nam YD, et al. PLoS One. 2011;6(7):e22109. doi: 10.1371/journal.pone.0022109. Epub 2011 Jul 29. PLoS One. 2011. PMID: 21829445 Free PMC article. - Application of phylogenetic microarrays to interrogation of human microbiota.
Paliy O, Agans R. Paliy O, et al. FEMS Microbiol Ecol. 2012 Jan;79(1):2-11. doi: 10.1111/j.1574-6941.2011.01222.x. Epub 2011 Nov 9. FEMS Microbiol Ecol. 2012. PMID: 22092522 Free PMC article. Review. - Microbes inside--from diversity to function: the case of Akkermansia.
Belzer C, de Vos WM. Belzer C, et al. ISME J. 2012 Aug;6(8):1449-58. doi: 10.1038/ismej.2012.6. Epub 2012 Mar 22. ISME J. 2012. PMID: 22437156 Free PMC article. Review.
Cited by
- Methodological Considerations in Longitudinal Analyses of Microbiome Data: A Comprehensive Review.
Lyu R, Qu Y, Divaris K, Wu D. Lyu R, et al. Genes (Basel). 2023 Dec 28;15(1):0. doi: 10.3390/genes15010051. Genes (Basel). 2023. PMID: 38254941 Free PMC article. Review. - Gut microbiota composition in depressive disorder: a systematic review, meta-analysis, and meta-regression.
Gao M, Wang J, Liu P, Tu H, Zhang R, Zhang Y, Sun N, Zhang K. Gao M, et al. Transl Psychiatry. 2023 Dec 8;13(1):379. doi: 10.1038/s41398-023-02670-5. Transl Psychiatry. 2023. PMID: 38065935 Free PMC article. - Microbes Promote Amino Acid Harvest to Rescue Undernutrition in Drosophila.
Yamada R, Deshpande SA, Bruce KD, Mak EM, Ja WW. Yamada R, et al. Cell Rep. 2015 Feb 17;10(6):865-872. doi: 10.1016/j.celrep.2015.01.018. Epub 2015 Feb 13. Cell Rep. 2015. PMID: 25683709 Free PMC article. - From gut dysbiosis to altered brain function and mental illness: mechanisms and pathways.
Rogers GB, Keating DJ, Young RL, Wong ML, Licinio J, Wesselingh S. Rogers GB, et al. Mol Psychiatry. 2016 Jun;21(6):738-48. doi: 10.1038/mp.2016.50. Epub 2016 Apr 19. Mol Psychiatry. 2016. PMID: 27090305 Free PMC article. Review. - The gut mycobiome signatures in long-lived populations.
Pu L, Pang S, Mu W, Chen X, Zou Y, Wang Y, Ding Y, Yan Q, Huang Y, Chen X, Peng T, Luo W, Wang S. Pu L, et al. iScience. 2024 Jun 28;27(8):110412. doi: 10.1016/j.isci.2024.110412. eCollection 2024 Aug 16. iScience. 2024. PMID: 39081291 Free PMC article.
References
- Zoetendal EG, Rajilic-Stojanovic M, de Vos WM. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut. 2008;57:1605–1615. - PubMed
- O'Toole PW, Claesson MJ. Gut microbiota: Changes throughout the lifespan from infancy to elderly. Int Dairy J. 2010;20:281–291.
- Raes J, Foerstner KU, Bork P. Get the most out of your metagenome: Computational analysis of environmental sequence data. Curr Opin Microbiol. 2007;10:490–498. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous